Fig. 1: Development of sc-SPORT to probe RNA structures in single cells.
From: RNA structure profiling at single-cell resolution reveals new determinants of cell identity
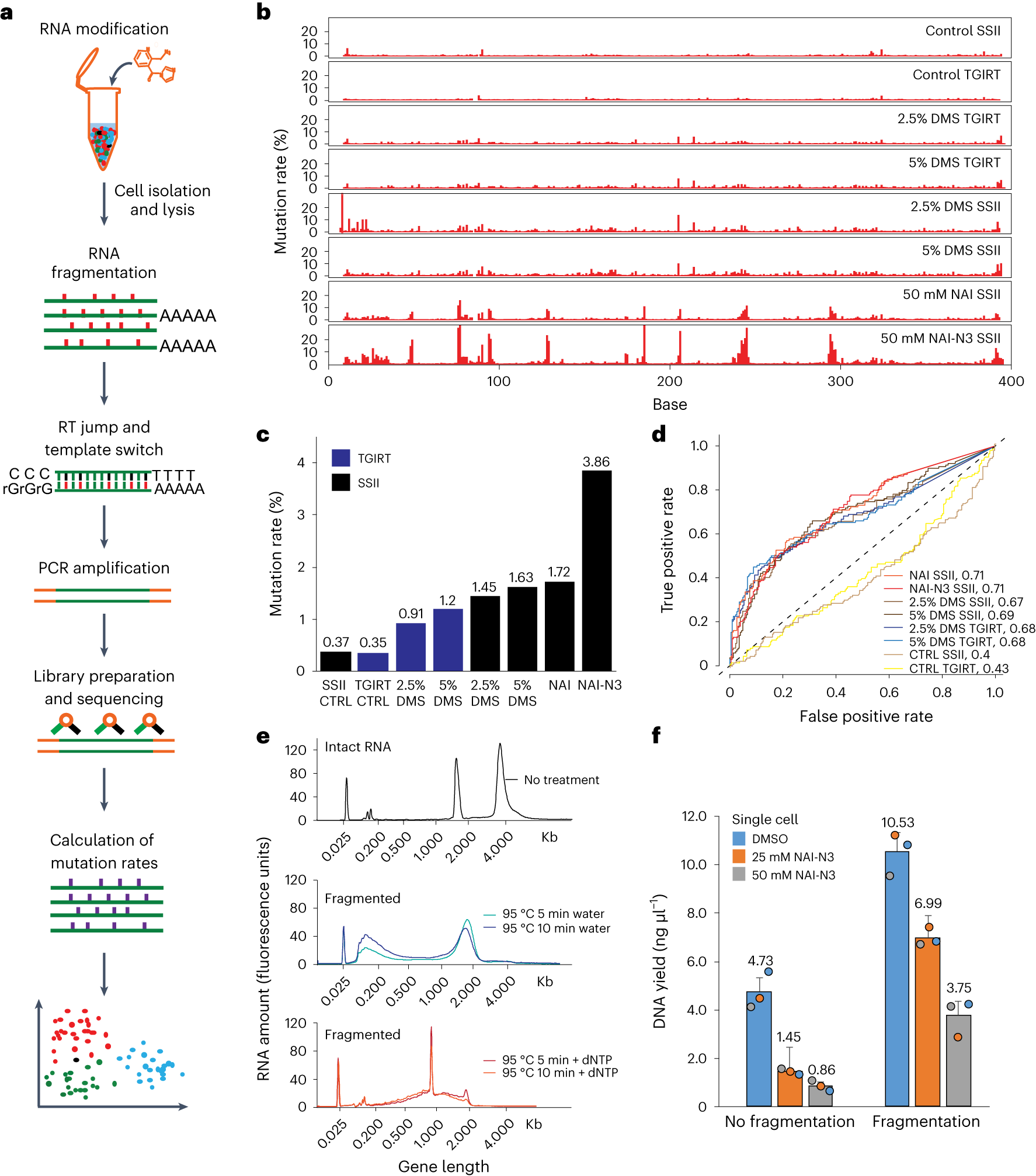
a, Experimental workflow of library preparation of sc-SPORT. b, Bar plots showing detected mutation rates on the Tetrahymena ribozyme using different RT enzymes (SSII and TGIRT) and different RNA modification compounds (DMS, NAI and NAI-N3) at different concentrations. c, A bar plot showing the average mutation rates along single-stranded regions of Tetrahymena ribozyme upon different treatments in b. d, ROC curves showing the accuracy of different treatments against the known secondary structure of Tetrahymena ribozyme. e, Bioanalyzer plots showing the size distribution of untreated RNA (top), RNA fragmented in water (middle) and RNA fragmented in water in the presence of 1 mM dNTP (bottom). f, Bar plots showing the amount of DNA that is generated from PCR amplification in single cells before (left) and after fragmentation (right) and in the presence of different concentrations of NAI-N3. The center represents the mean, and the error bars show the standard deviation. N = 3 biological replicates.
