Abstract
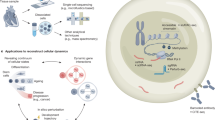
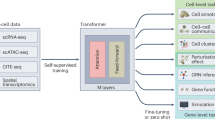
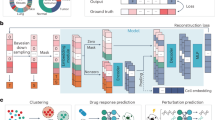
Transformer-based models are rapidly becoming foundational tools for analyzing and integrating multiscale biological data. This Perspective examines recent advances in transformer architectures, tracing their evolution from unimodal and augmented unimodal models to large-scale multimodal foundation models operating across genomic sequences, single-cell transcriptomics and spatial data. We categorize these models into three tiers and evaluate their capabilities for structural learning, representation transfer and tasks such as cell annotation, prediction and imputation. While discussing tokenization, interpretability and scalability challenges, we highlight emerging approaches that leverage masked modeling, contrastive learning and large language models. To support broader adoption, we provide practical guidance through code-based primers, using public datasets and open-source implementations. Finally, we propose designing a modular ‘Super Transformer’ architecture using cross-attention mechanisms to integrate heterogeneous modalities. This Perspective serves as a resource and roadmap for leveraging transformer models in multiscale, multimodal genomics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Code availability
Code for primers is available and can be run on Google Colab via https://colab.research.google.com/drive/16VxwUb3TQXulSGDdBW8gHG4elp8Rs92s/ (multi-omics primer), https://colab.research.google.com/drive/1YX_uO73lr8uENXLLj57cMHn796PtAoVd/ (genomic sequence analysis primer), https://colab.research.google.com/drive/1yDKEFXLIr884JeBDQMHWYthpa-u8k3q9/ (single-cell genomics primer) and https://colab.research.google.com/drive/13kax9iVi4uI6sh3ciXL9HxLl_RNtBcmy/ (spatial transcriptomics primer). The primers, as well as the associated data, are available on GitHub at https://github.com/TranslationalBioinformaticsUnit/Transformers-for-Multiscale-Genomics?tab=readme-ov-file/. The GitHub page for all models (papers) is available via https://github.com/TranslationalBioinformaticsUnit/TransformersInGenomicsPapers/.
References
Sapoval, N. et al. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 13, 1728 (2022).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015). Introduced DeepBind, a deep-learning model to infer protein–DNA/RNA binding motifs directly from sequence.
Zhou, J. & Troyanskaya, O. G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015).
Kelley, D. R., Snoek, J. & Rinn, J. L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–999 (2016).
Zou, J. et al. A primer on deep learning in genomics. Nat. Genet. 51, 12–18 (2019).
Eraslan, G., Avsec, Ž, Gagneur, J. & Theis, F. J. Deep learning: new computational modelling techniques for genomics. Nat. Rev. Genet. 20, 389–403 (2019).
Luecken, M. D. & Theis, F. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Vaswani, A. et al. Attention is all you need. In 31st Conference on Neural Information Processing Systems (eds von Luxburg, E. et al.) 6000–6010 (Curran Associates, 2017). Introduced the transformer architecture and self-attention mechanism, laying the algorithmic foundation for today’s foundation models.
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Vol. 1 (Long and Short Papers), 4171–4186 (2018).
OpenAI et al. GPT-4 Technical Report (OpenAI, 2023).
Dosovitskiy, A. et al. An image is worth 16×16 words: transformers for image recognition at scale. In International Conference on Learning Representations https://openreview.net/pdf?id=YicbFdNTTy (2020).
Consens, M. E. et al. Transformers and genome language models. Nat. Mach. Intell. 7, 346–362 (2025).
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://arxiv.org/abs/2108.07258 (2021).
Szałata, A. et al. Transformers in single-cell omics: a review and new perspectives. Nat. Methods 21, 1430–1443 (2024).
Greener, J. G., Kandathil, S. M., Moffat, L. & Jones, D. T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23, 40–55 (2022).
Simon, E., Swanson, K. & Zou, J. Language models for biological research: a primer. Nat. Methods 21, 1422–1429 (2024).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25, 24–29 (2019).
Cui, H. et al. Towards multimodal foundation models in molecular cell biology. Nature 640, 623–633 (2025).
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Regev, A., Teichmann, S. A. & Lander, E. S. The Human Cell Atlas. Elife https://doi.org/10.7554/eLife.27041.001 (2017). Laid out the initial roadmap for a comprehensive reference map of all human cell types, catalyzing a global single-cell genomics effort.
HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Huber, W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature https://doi.org/10.1038/s41586-021-03819-2 (2021). Demonstrated near-experimental accuracy in protein folding, marking a paradigm shift that validated deep learning as a tool for solving grand challenges in structural biology.
Radford, A. et al. Robust speech recognition via large-scale weak supervision. Proc. Mach. Learn. Res. 202, 28492–28518 (2022).
Tay, Y., Dehghani, M., Bahri, D. & Metzler, D. Efficient transformers: a survey. ACM Comput. Surv. 55, 1–28 (2022).
Bishop, C. M. & Bishop, H. Deep Learning: Foundations and Concepts (Springer, 2024).
Umarov, R. K. & Solovyev, V. V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE 12, e0171410 (2017).
Oubounyt, M., Louadi, Z., Tayara, H. & To Chong, K. DeePromoter: robust promoter predictor using deep learning. Front. Genet. 10, 286 (2019).
Zhu, Y. et al. Computational identification of eukaryotic promoters based on cascaded deep capsule neural networks. Brief Bioinform. 22, bbaa299 (2021).
Ma, Z. W., Zhao, J. P., Tian, J. & Zheng, C. H. DeeProPre: a promoter predictor based on deep learning. Comput. Biol. Chem. 101, 107770 (2022).
Choi, S. R. & Lee, M. Transformer architecture and attention mechanisms in genome data analysis: a comprehensive review. Biology 12, 1033 (2023).
Ji, Y., Zhou, Z., Liu, H. & Davuluri, R. V. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome. Bioinformatics 37, 2112–2120 (2021).
Li, Y. et al. msBERT-Promoter: a multi-scale ensemble predictor based on BERT pre-trained model for the two-stage prediction of DNA promoters and their strengths. BMC Biol. 22, 126 (2024).
Zhou, Z. et al. DNABERT-2: efficient foundation model and benchmark for multi-species genomes. In International Conference on Representation Learning (vol. 2024), 41642–41665 (2024).
Dalla-Torre, H. et al. Nucleotide transformer: building and evaluating robust foundation models for human genomics. Nat. Methods https://doi.org/10.1038/s41592-024-02523-z (2024).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. https://doi.org/10.1038/s41576-023-00586-w (2023).
Yang, F. et al. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. https://doi.org/10.1038/s42256-023-00757-8 (2022).
Khan, S. A. et al. Reusability report: learning the transcriptional grammar in single-cell RNA-sequencing data using transformers. Nat. Mach. Intell. https://doi.org/10.1038/s42256-023-00757-8 (2023).
Hao, M. et al. Large-scale foundation model on single-cell transcriptomics. Nat. Methods https://doi.org/10.1038/s41592-024-02305-7 (2024).
Gong, J. et al. xTrimoGene: an efficient and scalable representation learner for single-cell RNA-seq data. In Advances in Neural Information Processing Systems (Vol. 36), 69391–69403 (2023).
Hu, Y. et al. Benchmarking algorithms for single-cell multi-omics prediction and integration. Nat. Methods 21, 2182–2194 (2024).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). A method to map whole-transcriptome expression across intact tissue sections, establishing the foundation of spatial transcriptomics.
Fang, S. et al. Computational approaches and challenges in spatial transcriptomics. Genomics Proteomics Bioinformatics 21, 24–47 (2023).
Palla, G., Fischer, D. S., Regev, A. & Theis, F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. 40, 308–318 (2022).
Zahedi, R. et al. Deep learning in spatially resolved transcriptomics: a comprehensive technical view. Brief Bioinform. 25, bbae082 (2024).
Wang, Z., Li, X., Takada, H. & Nagatomi, R. stBERT: a pretrained model for spatial domain identification of spatial transcriptomics. IEEE Access https://doi.org/10.1109/ACCESS.2024.3479153 (2024).
Xu, B., Zheng, S. & Jin, Q. SPAFormer: sequential 3D part assembly with transformers. In 2025 International Conference on 3D Vision (3DV), Singapore, Singapore, 1317–1327 (2025).
Xue, S., Zhu, F., Wang, C. & Min, W. in Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 14954, 63–75 (Springer, 2024).
Cao, S. et al. stFormer: a foundation model for spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2024.09.27.615337 (2024).
Wiegreffe, S. & Pinter, Y. Attention is not not explanation. In EMNLP-IJCNLP 2019 - 2019 Conference on Empirical Methods in Natural Language Processing and 9th International Joint Conference on Natural Language Processing, Proceedings of the Conference 11–20 (2019).
Csendes, G., Szalay, K. Z. & Szalai, B. Benchmarking foundational cell models for post-perturbation RNA-seq prediction. BMC Genomics 26, 393 (2025).
Ahlmann-Eltze, C., Huber, W. & Anders, S. Deep-learning-based gene perturbation effect prediction does not yet outperform simple linear baselines. Nat. Methods 22, 1657–1661 (2025).
Alsabbagh, A. R. et al. Foundation models meet imbalanced single-cell data when learning cell type annotations. Preprint at bioRxiv https://doi.org/10.1101/2023.10.24.563625 (2023).
Kedzierska, K. Z. et al. Zero-shot evaluation reveals limitations of single-cell foundation models. Genome Biol. 26, 101 (2025).
Marin, F. I. et al. BEND: benchmarking DNA language models on biologically meaningful tasks. Preprint at https://arxiv.org/abs/2311.12570 (2024).
Bernett, J. et al. Guiding questions to avoid data leakage in biological machine learning applications. Nat. Methods 21, 1444–1453 (2024).
Avsec, Ž et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods 18, 1196–1203 (2021). Introducing Enformer, a transformer that models >200-kb genomic contexts end-to-end, setting a benchmark for sequence-to-expression prediction.
Chu, Y. et al. A 5′ UTR language model for decoding untranslated regions of mRNA and function predictions. Nat. Mach. Intell. 6, 449–460 (2024).
Mendoza-Revilla, J. et al. A foundational large language model for edible plant genomes. Commun. Biol. 7, 835 (2024).
Nguyen, E. et al. Sequence modeling and design from molecular to genome scale with Evo. Science 386, eado9336 (2024).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at https://arxiv.org/abs/2001.08361 (2020).
Almotairi, S., Badr, E., Abdelbaky, I., Elhakeem, M. & Abdul Salam, M. Hybrid transformer-CNN model for accurate prediction of peptide hemolytic potential. Sci. Rep. 14, 14263 (2024).
Shao, R., Bi, X. J. & Chen, Z. A novel hybrid transformer-CNN architecture for environmental microorganism classification. PLoS ONE 17, e0277557 (2022).
Tharani Pavithra, P. & Baranidharan, B. A hybrid ViT-CNN model premeditated for rice leaf disease identification. Int. J. Comput. Methods Exp. Meas. 12, 35–43 (2024).
Jia, Y., Liu, J., Chen, L., Zhao, T. & Wang, Y. THItoGene: a deep learning method for predicting spatial transcriptomics from histological images. Brief Bioinform. 25, bbad464 (2024).
Zeng, Y. et al. Spatial transcriptomics prediction from histology jointly through Transformer and graph neural networks. Brief Bioinform. 23, bbac297 (2022).
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods https://doi.org/10.1038/s41592-024-02201-0 (2024). A generative pretrained transformer model leveraging 33 million cells, illustrating the power of GPT-style models to advance single-cell biology.
Wang, X. et al. MarsGT: Multi-omics analysis for rare population inference using single-cell graph transformer. Nat. Commun. 15, 338 (2024).
Xiong, L., Chen, T. & Kellis, M. scCLIP: multi-modal single-cell contrastive learning integration pre-training. In NeurIPS AI for Science Workshop 2023 https://openreview.net/forum?id=KMtM5ZHxct (2023).
Li, G. et al. A deep generative model for multi-view profiling of single-cell RNA-seq and ATAC-seq data. Genome Biol. 23, 20 (2022).
Ma, A. et al. Single-cell biological network inference using a heterogeneous graph transformer. Nat. Commun. 14, 964 (2023).
He, Y. et al. Generalized biological foundation model with unified nucleic acid and protein language. Nat. Mach. Intell. 7, 942–953 (2025).
Li, X., Zhu, F. & Min, W. SpaDiT: diffusion transformer for spatial gene expression prediction using scRNA-seq. Brief Bioinform. https://doi.org/10.1093/bib/bbae571 (2024).
Tejada-Lapuerta, A. et al. Nicheformer: a foundation model for single-cell and spatial omics. Nat. Methods https://doi.org/10.1038/s41592-025-02814-z (2025).
Xu, J., Huang, D. S. & Zhang, X. scmFormer integrates large-scale single-cell proteomics and transcriptomics data by multi-task transformer. Adv. Sci. 11, e2307835 (2024).
Choi, H. et al. CELLama: foundation model for single cell and spatial transcriptomics by cell embedding leveraging language model abilities. Preprint at bioRxiv https://doi.org/10.1101/2024.05.08.593094 (2024).
Wen, H. et al. CellPLM: pre-training of cell language model beyond single cells. in International Conference on Learning Representations https://openreview.net/pdf?id=BKXvPDekud (2024).
Li, C. et al. scInterpreter: training large language models to interpret scRNA-seq data for cell type annotation. Preprint at https://arxiv.org/abs/2402.12405 (2024).
Schaefer, M. et al. Multimodal learning of transcriptomes and text enables interactive single-cell RNA-seq data exploration with natural-language chats. Preprint at bioRxiv https://doi.org/10.1101/2024.10.15.618501 (2024).
de Almeida, B. P. et al. A multimodal conversational agent for DNA, RNA and protein tasks. Nat. Mach. Intell. 7, 928–941 (2025).
Lu, Y. -C. et al. scChat: a large language model-powered co-pilot for contextualized single-cell RNA sequencing analysis. Preprint at bioRxiv https://doi.org/10.1101/2024.10.01.616063 (2024).
Levine, D. et al. Cell2Sentence: teaching large language models the language of biology. Proc. Mach. Learn. Res. 235, 27299–27325 (2024).
Chen, Y. & Zou, J. Simple and effective embedding model for single-cell biology built from ChatGPT. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-024-01284-6 (2024). Demonstrated that ChatGPT-derived gene embeddings can be averaged into cell embeddings, enabling zero-shot single-cell analyses without large omics pretraining.
Terven, J., Cordova-Esparza, D. M., Ramirez-Pedraza, A., Chavez-Urbiola, E. A. & Romero-Gonzalez, J. A. Loss functions and metrics in deep learning. Preprint at https://arxiv.org/abs/2307.02694 (2023).
Radford, A. et al. Learning transferable visual models from natural language supervision. Proc. Mach. Learn. Res. 139, 8748–8763 (2021).
Jain, S. et al. CAGI, the Critical Assessment of Genome Interpretation, establishes progress and prospects for computational genetic variant interpretation methods. Genome Biol. 25, 53 (2024).
Choromanski, K. et al. Rethinking attention with performers. In International Conference on Learning Representations https://openreview.net/pdf?id=Ua6zuk0WRH (2021).
Wang, H. et al. Scientific discovery in the age of artificial intelligence. Nature 620, 47–60 (2023). An important perspective outlining how AI systems, from language models to multimodal transformers, are reshaping hypothesis generation, experimental design and scientific governance.
Beltagy, I., Peters, M. E. & Cohan, A. Longformer: the long-document transformer. Preprint at https://arxiv.org/abs/2004.05150 (2020).
Kitaev, N., Kaiser, Ł. & Levskaya, A. Reformer: the efficient transformer. In International Conference on Learning Representations https://openreview.net/pdf?id=rkgNKkHtvB (2020).
Wang, S., Li, B. Z., Khabsa, M., Fang, H. & Ma, H. Linformer: self-attention with linear complexity. Preprint at https://arxiv.org/abs/2006.04768 (2020).
Badia-i-Mompel, P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24, 739–754 (2023).
Armingol, E., Baghdassarian, H. M. & Lewis, N. E. The diversification of methods for studying cell–cell interactions and communication. Nat. Rev. Genet. 25, 381–400 (2024).
Shen, T. et al. Accurate RNA 3D structure prediction using a language model-based deep learning approach. Nat. Methods https://doi.org/10.1038/s41592-024-02487-0 (2024).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 2023 (2023).
Tegnér, J. N. et al. Computational disease modeling - fact or fiction? BMC Syst. Biol. 3, 56 (2009).
Bunne, C. et al. How to build the virtual cell with artificial intelligence: priorities and opportunities. Cell 187, 7045–7063 (2024).
Lagani, V., Triantafillou, S., Ball, G., Tegnér, J. & Tsamardinos, I. in Uncertainty in Biology (eds Geris, L. & Gomez-Cabrero, D.) 33–73 (Springer, 2016).
Acknowledgements
King Abdullah University of Science and Technology supported this work. We thank all members of the laboratory for constructive, critical reading.
Author information
Authors and Affiliations
Contributions
S.A.K., X.M.-d.-M., R.L. and J.T. conceptualized the study. S.A.K., X.M.-d.-M., R.L. and J.T. drafted the initial manuscript. S.A.K., X.M.-d.-M., R.L., A.R.A., V.L., D.G.-C. and J.T. refined the concepts and methodology. A.R.A. and R.L. prepared the figures. J.T. supervised the overall research direction and advised on critical revisions. All authors reviewed, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Hani Goodarzi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–4, Boxes 1–3, Fig. 1 and Tables 1 and 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khan, S.A., Martínez-de-Morentin, X., Alsabbagh, A.R. et al. Multimodal foundation transformer models for multiscale genomics. Nat Methods (2025). https://doi.org/10.1038/s41592-025-02918-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41592-025-02918-6