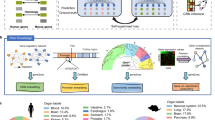
Abstract
Research organisms provide invaluable insights into human biology and diseases, serving as essential tools for functional experiments, disease modeling and drug testing. However, evolutionary divergence between humans and research organisms hinders effective knowledge transfer across species. Here, we review state-of-the-art methods for computationally transferring knowledge across species, primarily focusing on methods that use transcriptome data and/or molecular networks. Our Perspective addresses four key areas: (1) transferring disease and gene annotation knowledge across species, (2) identifying functionally equivalent molecular components, (3) inferring equivalent perturbed genes or gene sets and (4) identifying equivalent cell types. We conclude with an outlook on future directions and several key challenges that remain in cross-species knowledge transfer, including introducing the concept of ‘agnology’ to describe functional equivalence of biological entities, regardless of their evolutionary origins. This concept is becoming pervasive in integrative data-driven models in which evolutionary origins of functions can remain unresolved.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Barré-Sinoussi, F. & Montagutelli, X. Animal models are essential to biological research: issues and perspectives. Future Sci. OA 1, FSO63 (2015).
Baldridge, D. et al. Model organisms contribute to diagnosis and discovery in the Undiagnosed Diseases Network: current state and a future vision. Orphanet J. Rare Dis. 16, 206 (2021).
Baumans, V. Use of animals in experimental research: an ethical dilemma? Gene Ther. 11, S64–S66 (2004).
Festing, S. & Wilkinson, R. The ethics of animal research. EMBO Rep. 8, 526–530 (2007).
Demers, G. et al. Harmonization of animal care and use guidance. Science 312, 700–701 (2006).
Wangler, M. F. et al. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207, 9–27 (2017).
Aitman, T. J. et al. The future of model organisms in human disease research. Nat. Rev. Genet. 12, 575–582 (2011).
McClellan, J. & King, M.-C. Genetic heterogeneity in human disease. Cell 141, 210–217 (2010).
Tadano, R. et al. Molecular characterization reveals genetic uniformity in experimental chicken resources. Exp. Anim. 59, 511–514 (2010).
Franěk, R. et al. Isogenic lines in fish — a critical review. Rev. Aquacult. 12, 1412–1434 (2019).
Cui, C., Zhou, Y. & Cui, Q. Defining the functional divergence of orthologous genes between human and mouse in the context of miRNA regulation. Brief. Bioinform. 22, bbab253 (2021).
Nehrt, N. L., Clark, W. T., Radivojac, P. & Hahn, M. W. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput. Biol. 7, e1002073 (2011).
Stamboulian, M., Guerrero, R. F., Hahn, M. W. & Radivojac, P. The ortholog conjecture revisited: the value of orthologs and paralogs in function prediction. Bioinformatics 36, i219–i226 (2020).
Nadimpalli, S., Persikov, A. V. & Singh, M. Pervasive variation of transcription factor orthologs contributes to regulatory network evolution. PLoS Genet. 11, e1005011 (2015).
Han, S. K., Kim, D., Lee, H., Kim, I. & Kim, S. Divergence of noncoding regulatory elements explains gene–phenotype differences between human and mouse orthologous genes. Mol. Biol. Evol. 35, 1653–1667 (2018).
Arendt, D. et al. The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016).
Büscher, T., Ganai, N., Gompper, G. & Elgeti, J. Tissue evolution: mechanical interplay of adhesion, pressure, and heterogeneity. New J. Phys. 22, 033048 (2020).
Ha, D. et al. Evolutionary rewiring of regulatory networks contributes to phenotypic differences between human and mouse orthologous genes. Nucleic Acids Res. 50, 1849–1863 (2022).
Rhrissorrakrai, K. et al. Understanding the limits of animal models as predictors of human biology: lessons learned from the sbv IMPROVER Species Translation Challenge. Bioinformatics 31, 471–483 (2014).
Zhou, N. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 20, 244 (2019).
Jiang, Y. et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biol. 17, 184 (2016).
Radivojac, P. et al. A large-scale evaluation of computational protein function prediction. Nat. Methods 10, 221–227 (2013).
Friedberg, I. & Radivojac, P. Community-wide evaluation of computational function prediction. In Methods in Molecular Biology 133–146 (Springer, 2016).
Brubaker, D. K. & Lauffenburger, D. A. Translating preclinical models to humans. Science 367, 742–743 (2020).
McWhite, C. D., Liebeskind, B. J. & Marcotte, E. M. Applications of comparative evolution to human disease genetics. Curr. Opin. Genet. Dev. 35, 16–24 (2015).
Kowald, A. et al. Transfer learning of clinical outcomes from preclinical molecular data, principles and perspectives. Brief. Bioinform. 23, bbac133 (2022).
Dougherty, B. V. & Papin, J. A. Systems biology approaches help to facilitate interpretation of cross-species comparisons. Curr. Opin. Toxicol. 23–24, 74–79 (2020).
Feuermann, M. et al. A compendium of human gene functions derived from evolutionary modelling. Nature 640, 146–154 (2025).
Wu, R. S. et al. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112–125 (2018).
D’Agostino, Y. et al. Loss of circadian rhythmicity in bdnf knockout zebrafish larvae. iScience 25, 104054 (2022).
Shearin, A. L. & Ostrander, E. A. Leading the way: canine models of genomics and disease. Dis. Model. Mech. 3, 27–34 (2010).
Castoe, T. A. et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl Acad. Sci. USA 110, 20645–20650 (2013).
Braasch, I. et al. The spotted gar genome illuminates vertebrate evolution and facilitates human–teleost comparisons. Nat. Genet. 48, 427–437 (2016).
Grohme, M. A. et al. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554, 56–61 (2018).
Warde-Farley, D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220 (2010).
Liu, R., Mancuso, C. A., Yannakopoulos, A., Johnson, K. A. & Krishnan, A. Supervised learning is an accurate method for network-based gene classification. Bioinformatics 36, 3457–3465 (2020).
Wang, X., Gulbahce, N. & Yu, H. Network-based methods for human disease gene prediction. Brief. Funct. Genomics 10, 280–293 (2011).
Singh-Blom, U. M. et al. Prediction and validation of gene–disease associations using methods inspired by social network analyses. PLoS ONE 8, e58977 (2013).
Yao, V. et al. An integrative tissue-network approach to identify and test human disease genes. Nat. Biotechnol. 36, 1091–1099 (2018). This study introduced diseaseQUEST, a method to combine human GWAS-derived gene–disease associations and research organism functional networks to predict candidate disease-related genes in research organisms.
Chikina, M. D. & Troyanskaya, O. G. Accurate quantification of functional analogy among close homologs. PLoS Comput. Biol. 7, e1001074 (2011). This method uses functional gene networks and metagenes of neighboring genes in these networks to find agnologs across species.
Singh, R., Xu, J. & Berger, B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl Acad. Sci. USA 105, 12763–12768 (2008).
Park, C. Y. et al. Functional knowledge transfer for high-accuracy prediction of under-studied biological processes. PLoS Comput. Biol. 9, e1002957 (2013). This study introduced the FKT method, a network-based approach that uses functional equivalents (that we call ‘agnologs’) to enhance gene annotation across species.
Yamaguchi, M., Imai, F., Tonou-Fujimori, N. & Masai, I. Mutations in N-cadherin and a Stardust homolog, Nagie oko, affect cell-cycle exit in zebrafish retina. Mech. Dev. 127, 247–264 (2010).
Mancuso, C. A., Johnson, K. A., Liu, R. & Krishnan, A. Joint representation of molecular networks from multiple species improves gene classification. PLoS Comput. Biol. 20, e1011773 (2024). This study introduced GenePlexusZoo, a method to simultaneously integrate network information across more than two species to improve gene classification.
Barot, M., Gligorijević, V., Cho, K. & Bonneau, R. NetQuilt: deep multispecies network-based protein function prediction using homology-informed network similarity. Bioinformatics 37, 2414–2422 (2021).
Katz, L. A new status index derived from sociometric analysis. Psychometrika 18, 39–43 (1953).
Liben-Nowell, D. & Kleinberg, J. The link‐prediction problem for social networks. J. Am. Soc. Inf. Sci. 58, 1019–1031 (2007).
Gligorijevic, D. Large-scale discovery of disease–disease and disease–gene associations. Sci. Rep. 6, 32404 (2016).
Sonawane, A. R. et al. Understanding tissue-specific gene regulation. Cell Rep. 21, 1077–1088 (2017).
Hekselman, I. & Yeger-Lotem, E. Mechanisms of tissue and cell-type specificity in heritable traits and diseases. Nat. Rev. Genet. 21, 137–150 (2020).
Greene, C. S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
Cowen, L., Ideker, T., Raphael, B. J. & Sharan, R. Network propagation: a universal amplifier of genetic associations. Nat. Rev. Genet. 18, 551–562 (2017).
Picart-Armada, S. et al. Benchmarking network propagation methods for disease gene identification. PLoS Comput. Biol. 15, e1007276 (2019).
Krishnan, A. et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci. 19, 1454–1462 (2016).
Mastropietro, A., De Carlo, G. & Anagnostopoulos, A. XGDAG: explainable gene–disease associations via graph neural networks. Bioinformatics 39, btad482 (2023).
Domazet-Loso, T. & Tautz, D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol. Biol. Evol. 25, 2699–2707 (2008).
Cai, J. J., Borenstein, E., Chen, R. & Petrov, D. A. Similarly strong purifying selection acts on human disease genes of all evolutionary ages. Genome Biol. Evol. 1, 131–144 (2009).
Lopez-Bigas, N. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 32, 3108–3114 (2004).
Dickerson, J. E. & Robertson, D. L. On the origins of Mendelian disease genes in man: the impact of gene duplication. Mol. Biol. Evol. 29, 61–69 (2011).
Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA 96, 4285–4288 (1999).
Marcotte, E. M. et al. Detecting protein function and protein–protein interactions from genome sequences. Science 285, 751–753 (1999).
Maxwell, E. K. et al. Evolutionary profiling reveals the heterogeneous origins of classes of human disease genes: implications for modeling disease genetics in animals. BMC Evol. Biol. 14, 212 (2014).
Amberger, J. S., Bocchini, C. A., Scott, A. F. & Hamosh, A. OMIM.org: leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res. 47, D1038–D1043 (2018).
Tabach, Y. et al. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol. Syst. Biol. 9, 692 (2013). This study uses phylogenetic profiles to enable systematic identifications of genome-wide functional modules that co-occur across the eukaryotic phylogeny.
Dey, G., Jaimovich, A., Collins, S. R., Seki, A. & Meyer, T. Systematic discovery of human gene function and principles of modular organization through phylogenetic profiling. Cell Rep. 10, 993–1006 (2015).
Brown, S. D. M. et al. High-throughput mouse phenomics for characterizing mammalian gene function. Nat. Rev. Genet. 19, 357–370 (2018).
Peterson, K. A. & Murray, S. A. Progress towards completing the mutant mouse null resource. Mamm. Genome 33, 123–134 (2021).
Alliance of Genome Resources Consortium. Harmonizing model organism data in the Alliance of Genome Resources. Genetics 220, iyac022 (2022).
Matentzoglu, N. et al. The Unified Phenotype Ontology : a framework for cross-species integrative phenomics. Genetics 229, iyaf027 (2025).
Hoehndorf, R., Schofield, P. N. & Gkoutos, G. V. PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res. 39, e119 (2011).
Smedley, D. et al. Next-generation diagnostics and disease–gene discovery with the Exomiser. Nat. Protoc. 10, 2004–2015 (2015).
Alghamdi, S. M., Schofield, P. N. & Hoehndorf, R. Contribution of model organism phenotypes to the computational identification of human disease genes. Dis. Model. Mech. 15, dmm049441 (2022). This study uses phenotype ontologies to relate research organism-specific and human-specific ontologies. By treating connected ontologies as graphs, they also proposed using graph embedding-based ML approaches to predict disease–gene associations across species.
Althagafi, A., Zhapa-Camacho, F. & Hoehndorf, R. Prioritizing genomic variants through neuro-symbolic, knowledge-enhanced learning. Bioinformatics 40, btae301 (2024).
Gherardini, P. F., Wass, M. N., Helmer-Citterich, M. & Sternberg, M. J. E. Convergent evolution of enzyme active sites is not a rare phenomenon. J. Mol. Biol. 372, 817–845 (2007).
Tulipano, A., Donvito, G., Licciulli, F., Maggi, G. & Gisel, A. Gene analogue finder: a GRID solution for finding functionally analogous gene products. BMC Bioinformatics 8, 329 (2007).
Li, H. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572–D580 (2006).
Fan, J. et al. Functional protein representations from biological networks enable diverse cross-species inference. Nucleic Acids Res. 47, e51 (2019). This study introduces MUNK, a method that uses aligned protein–protein interaction network representations to identify agnologous gene pairs. Additionally, this work expands the concept of phenologs, characterizing them by a significant overlap of agnologous gene pairs.
Arsenescu, V. et al. MUNDO: protein function prediction embedded in a multispecies world. Bioinform. Adv. 2, vbab025 (2021).
Li, L. et al. Joint embedding of biological networks for cross-species functional alignment. Bioinformatics 39, btad529 (2023).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Rives, A. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021).
Hamamsy, T. et al. Protein remote homology detection and structural alignment using deep learning. Nat. Biotechnol. 42, 975–985 (2023).
Hong, L. et al. Fast, sensitive detection of protein homologs using deep dense retrieval. Nat. Biotechnol. 43, 983–995 (2025).
Brixi, G. et al. Genome modeling and design across all domains of life with Evo 2. Preprint at bioRxiv https://doi.org/10.1101/2025.02.18.638918 (2025).
McGary, K. L. et al. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl Acad. Sci. USA 107, 6544–6549 (2010). This study introduced the concept of phenologs, which facilitates finding phenotypic counterparts across species by looking at ortholog overlap between genes annotated to phenotypes in different species.
Lee, J., Shah, M., Ballouz, S., Crow, M. & Gillis, J. CoCoCoNet: conserved and comparative co-expression across a diverse set of species. Nucleic Acids Res. 48, W566–W571 (2020). This study introduced CoCoCoNet, a method that helps to find agnologous gene sets across species by comparing coexpression networks generated in different species.
El-Kebir, M. et al. xHeinz: an algorithm for mining cross-species network modules under a flexible conservation model. Bioinformatics 31, 3147–3155 (2015).
Deshpande, R., Sharma, S., Verfaillie, C. M., Hu, W.-S. & Myers, C. L. A scalable approach for discovering conserved active subnetworks across species. PLoS Comput. Biol. 6, e1001028 (2010). This study introduced neXus, a method that integrates DEG lists and network topology to find agnologous gene modules across species.
Zinman, G. E. et al. ModuleBlast: identifying activated sub-networks within and across species. Nucleic Acids Res. 43, e20 (2014).
Zong, W. et al. Transcriptomic congruence analysis for evaluating model organisms. Proc. Natl Acad. Sci. USA 120, e2202584120 (2023). This study introduced CAMO, a method to systematically quantify the similarity between perturbed transcriptomic profiles across species.
Seok, J. et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA 110, 3507–3512 (2013).
Takao, K. & Miyakawa, T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA 112, 1167–1172 (2014).
Putman, T. E. et al. The Monarch Initiative in 2024: an analytic platform integrating phenotypes, genes and diseases across species. Nucleic Acids Res. 52, D938–D949 (2023).
Normand, R. et al. Found in Translation: a machine learning model for mouse-to-human inference. Nat. Methods 15, 1067–1073 (2018). This study introduced FIT, a method that investigated the possibility of transferring DEG results across species using linear models.
Brubaker, D. K., Proctor, E. A., Haigis, K. M. & Lauffenburger, D. A. Computational translation of genomic responses from experimental model systems to humans. PLoS Comput. Biol. 15, e1006286 (2019).
Brubaker, D. K. et al. An interspecies translation model implicates integrin signaling in infliximab-resistant inflammatory bowel disease. Sci. Signal. 13, eaay3258 (2020). This study introduced TransComp-R, a method that allows transferring knowledge across species when the data from each species were not measured using the same omics modality.
Suarez-Lopez, L. et al. Cross-species transcriptomic signatures predict response to MK2 inhibition in mouse models of chronic inflammation. iScience 24, 103406 (2021).
Lee, M. J. et al. Computational interspecies translation between Alzheimer’s disease mouse models and human subjects identifies innate immune complement, TYROBP, and TAM receptor agonist signatures. Front. Neurosci. 15, 727784 (2021).
Carroll, M. J., Garcia-Reyero, N., Perkins, E. J. & Lauffenburger, D. A. Translatable pathways classification (TransPath-C) for inferring processes germane to human biology from animal studies data: example application in neurobiology. Integr. Biol. 13, 237–245 (2021).
Russkikh, N. et al. Style transfer with variational autoencoders is a promising approach to RNA-seq data harmonization and analysis. Bioinformatics 36, 5076–5085 (2020).
Lotfollahi, M., Wolf, F. A. & Theis, F. J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Meimetis, N. et al. AutoTransOP: translating omics signatures without orthologue requirements using deep learning. NPJ Syst. Biol. Appl. 10, 13 (2024).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Cai, M., Hao Nguyen, C., Mamitsuka, H. & Li, L. XGSEA: CROSS-species gene set enrichment analysis via domain adaptation. Brief. Bioinform. 22, bbaa406 (2021).
Ali, A. et al. The ancestral gene repertoire of animal stem cells. Proc. Natl Acad. Sci. USA 112, E7093–E7100 (2015).
Shafer, M. E. R. Cross-species analysis of single-cell transcriptomic data. Front. Cell Dev. Biol. 7, 175 (2019).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2021).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887 (2019).
Bailon-Zambrano, R. et al. Variable paralog expression underlies phenotype variation. eLife 11, e79247 (2022).
Forslund, K., Schreiber, F., Thanintorn, N. & Sonnhammer, E. L. L. OrthoDisease: tracking disease gene orthologs across 100 species. Brief. Bioinform. 12, 463–473 (2011).
Song, Y., Miao, Z., Brazma, A. & Papatheodorou, I. Benchmarking strategies for cross-species integration of single-cell RNA sequencing data. Nat. Commun. 14, 6495 (2023).
Tarashansky, A. J. et al. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10, e66747 (2021). This study introduced SAMap, a method that fully uses complex homology patterns to align single-cell transcriptomes across a pair of species.
Rosen, Y. et al. Toward universal cell embeddings: integrating single-cell RNA-seq datasets across species with SATURN. Nat. Methods 21, 1492–1500 (2024). This study introduced SATURN, a method that uses all genes, regardless of their evolutionary relationships, to align single-cell transcriptomes across species. SATURN is also able to simultaneously integrate single-cell transcriptomes from more than two species.
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Galkin, F. et al. Precious3GPT: multimodal multi-species multi-omics multi-tissue transformer for aging research and drug discovery. Preprint at bioRxiv https://doi.org/10.1101/2024.07.25.605062 (2024).
Yang, X. et al. GeneCompass: deciphering universal gene regulatory mechanisms with a knowledge-informed cross-species foundation model. Cell Res. 34, 830–845 (2024). This method employs a transformer-based architecture to align agnologous cells across species while integrating auxiliary information such as gene regulatory networks, gene promoters, gene families and coexpression networks.
Rosen, Y. et al. Universal Cell Embeddings: a foundation model for cell biology. Preprint at bioRxiv https://doi.org/10.1101/2023.11.28.568918 (2023).
Marian, A. J. Modeling human disease phenotype in model organisms. Circ. Res. 109, 356–359 (2011).
Li, Y. & Agarwal, P. A pathway-based view of human diseases and disease relationships. PLoS ONE 4, e4346 (2009).
Cui, P., Ma, X., Li, H., Lang, W. & Hao, J. Shared biological pathways between Alzheimer’s disease and ischemic stroke. Front. Neurosci. 12, 605 (2018).
Gokuladhas, S., Schierding, W., Golovina, E., Fadason, T. & O'Sullivan, J. Unravelling the shared genetic mechanisms underlying 18 autoimmune diseases using a systems approach. Front. Immunol. 12, 693142 (2021).
Tokarek, J. et al. Molecular processes involved in the shared pathways between cardiovascular diseases and diabetes. Biomedicines 11, 2611 (2023).
Stoney, R., Robertson, D. L., Nenadic, G. & Schwartz, J.-M. Mapping biological process relationships and disease perturbations within a pathway network. NPJ Syst. Biol. Appl. 4, 22 (2018).
Hickey, S. L., McKim, A., Mancuso, C. A. & Krishnan, A. A network-based approach for isolating the chronic inflammation gene signatures underlying complex diseases towards finding new treatment opportunities. Front. Pharmacol. 13, 995459 (2022).
Buphamalai, P., Kokotovic, T., Nagy, V. & Menche, J. Network analysis reveals rare disease signatures across multiple levels of biological organization. Nat. Commun. 12, 6306 (2021).
Gargano, M. A. et al. The Human Phenotype Ontology in 2024: phenotypes around the world. Nucleic Acids Res. 52, D1333–D1346 (2023).
Blair, D. R., Hoffmann, T. J. & Shieh, J. T. Common genetic variation associated with Mendelian disease severity revealed through cryptic phenotype analysis. Nat. Commun. 13, 3675 (2022).
Maassen, W. et al. Curation and expansion of the Human Phenotype Ontology for systemic autoinflammatory diseases improves phenotype-driven disease-matching. Front. Immunol. 14, 1215869 (2023).
Brunet, F. G. et al. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 23, 1808–1816 (2006).
True, J. R. & Haag, E. S. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3, 109–119 (2001).
Ghadie, M. A., Coulombe-Huntington, J. & Xia, Y. Interactome evolution: insights from genome-wide analyses of protein–protein interactions. Curr. Opin. Struct. Biol. 50, 42–48 (2018).
Ecovoiu, A. A., Ratiu, A. C., Micheu, M. M. & Chifiriuc, M. C. Inter-species rescue of mutant phenotype—the standard for genetic analysis of human genetic disorders in Drosophila melanogaster model. Int. J. Mol. Sci. 23, 2613 (2022).
Kachroo, A. H. et al. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921–925 (2015).
Futuyma, D. J. Evolution (Sinauer Associates, 2013).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2022).
Szymborski, J. & Emad, A. INTREPPPID—an orthologue-informed quintuplet network for cross-species prediction of protein–protein interaction. Brief. Bioinform. https://doi.org/10.1093/bib/bbae405 (2024).
Walhout, A. J. M. et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287, 116–122 (2000).
Yu, H. et al. Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res. 14, 1107–1118 (2004).
Wong, A. K., Krishnan, A., Yao, V., Tadych, A. & Troyanskaya, O. G. IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 43, W128–W133 (2015).
Sealfon, R. S. G., Wong, A. K. & Troyanskaya, O. G. Machine learning methods to model multicellular complexity and tissue specificity. Nat. Rev. Mater. 6, 717–729 (2021).
Saha, A. et al. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 27, 1843–1858 (2017).
Burns, J. J. R. et al. Addressing noise in co-expression network construction. Brief. Bioinform. 23, bbab495 (2021).
Xue, S. et al. Applying differential network analysis to longitudinal gene expression in response to perturbations. Front. Genet. 13, 1026487 (2022).
Liu, R., Yuan, H., Johnson, K. & Krishnan, A. CONE: context-specific network embedding via contextualized graph attention. In Proc. 19th Machine Learning in Computational Biology Meeting (eds. Knowles, D. A. & Mostafavi, S.) 261, 53–71 (2024).
Li, M. M. et al. Contextual AI models for single-cell protein biology. Nat. Methods 21, 1546–1557 (2024).
Zitnik, M. & Leskovec, J. Predicting multicellular function through multi-layer tissue networks. Bioinformatics 33, i190–i198 (2017).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2021).
Zhou, J. et al. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet. 50, 1171–1179 (2018).
Sokolova, K. et al. Atlas of primary cell-type-specific sequence models of gene expression and variant effects. Cell Rep. Methods 3, 100580 (2023).
Rhie, A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021).
Li, M. M., Huang, K. & Zitnik, M. Graph representation learning in biomedicine and healthcare. Nat. Biomed. Eng. 6, 1353–1369 (2022).
Bradshaw, M. S., Gaskell, A. & Layer, R. M. The effects of biological knowledge graph topology on embedding-based link prediction. Preprint at bioRxiv https://doi.org/10.1101/2024.06.10.598277 (2024).
O’Neil, S. T. et al. Phenomics Assistant: an interface for LLM-based biomedical knowledge graph exploration. Preprint at bioRxiv https://doi.org/10.1101/2024.01.31.578275 (2024).
Gaudet, P. & Dessimoz, C. Gene Ontology: pitfalls, biases, and remedies. In Methods in Molecular Biology 189–205 (Springer, 2016).
Caufield, J. H. et al. Structured Prompt Interrogation and Recursive Extraction of Semantics (SPIRES): a method for populating knowledge bases using zero-shot learning. Bioinformatics https://doi.org/10.1093/bioinformatics/btae104 (2024).
Howe, K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013).
Russell, J. J. et al. Non-model model organisms. BMC Biol. 15, 55 (2017).
Smith, J. J. et al. A chromosome-scale assembly of the axolotl genome. Genome Res. 29, 317–324 (2019).
Kourakis, M. J. & Smith, W. C. An organismal perspective on C. intestinalis development, origins and diversification. eLife 4, e06024 (2015).
Acknowledgements
This work is supported by NIH R35 GM128765 and Simons Foundation 1017799 (to A.K.). (Zebra)fish–human research transfer in the Braasch laboratory has been supported by NIH R01 OD011116. We thank all members in the Krishnan and Braasch laboratories for helpful discussion and feedback on the manuscript.
Author information
Authors and Affiliations
Contributions
H.Y. drafted the manuscript. H.Y., C.A.M., K.J., I.B. and A.K. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Ran Ran, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Madhura Mukhopadhyay, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note 1
Supplementary Table 1
Summary of reviewed methods.
Supplementary Table 2
Data related to reviewed methods.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, H., Mancuso, C.A., Johnson, K. et al. Computational strategies for cross-species knowledge transfer. Nat Methods (2025). https://doi.org/10.1038/s41592-025-02931-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41592-025-02931-9