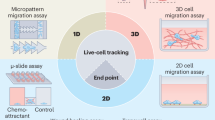
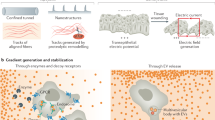
Abstract
Cell migration assays provide invaluable insights into fundamental biological processes. In a companion Review, we describe commercial and custom in vitro and in vivo assays to measure cell migration and provide guidelines on how to select the most appropriate assay for a given biological question. Here, we describe the fundamental principles of how to compute—from the raw data generated by these assays—quantitative cell migration parameters that help determine the biophysical nature of the cell migration, such as cell speed, mean-squared displacement, diffusivity, persistence, speed and anisotropy, and how to quantify cell heterogeneity, with practical guidance. We also describe new imaging and computational technologies, including AI-based methods, which have helped establish fast, robust and accurate tracking of cells and quantification of cell migration. Taken together, these Reviews offer practical guidance for cell migration assays from conception to analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Luster, A. D., Alon, R. & von Andrian, U. H. Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 6, 1182–1190 (2005.
Fowell, D. J. & Kim, M. The spatio-temporal control of effector T cell migration. Nat. Rev. Immunol. 21, 582–596 (2021).
Horwitz, R. & Webb, D. Cell migration. Curr. Biol. 13, R756–R759 (2003).
Scarpa, E. & Mayor, R. Collective cell migration in development. J. Cell Biol. 212, 143–155 (2016).
Kurosaka, S. & Kashina, A. Cell biology of embryonic migration. Birth Defects Res. C. Embryo Today 84, 102–122 (2008).
Gurtner, G. C., Werner, S., Barrandon, Y. & Longaker, M. T. Wound repair and regeneration. Nature 453, 314–321 (2008).
Arwert, E. N., Hoste, E. & Watt, F. M. Epithelial stem cells, wound healing and cancer. Nat. Rev. Cancer 12, 170–180 (2012).
Coulombe, P. A. Wound epithelialization: accelerating the pace of discovery. J. Invest. Dermatol. 121, 219–230 (2003).
Aragona, M. et al. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 8, 14684 (2017).
Friedl, P. & Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374 (2003).
Yamaguchi, H., Wyckoff, J. & Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 17, 559–564 (2005).
Castaneda, M., den Hollander, P., Kuburich, N. A., Rosen, J. M. & Mani, S. A. Mechanisms of cancer metastasis. Semin. Cancer Biol. 87, 17–31 (2022).
Popper, H. H. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 35, 75–91 (2016).
Jones, D. H. et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature 440, 692–696 (2006).
Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 (2019).
Fares, J., Fares, M. Y., Khachfe, H. H., Salhab, H. A. & Fares, Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target. Ther. 5, 28 (2020).
Mackay, C. R. Moving targets: cell migration inhibitors as new anti-inflammatory therapies. Nat. Immunol. 9, 988–998 (2008).
Min, H. Y. & Lee, H. Y. Molecular targeted therapy for anticancer treatment. Exp. Mol. Med. 54, 1670–1694 (2022).
Du, W. et al. Selecting the optimal cell migration assay: fundamentals and practical guidelines. Nat. Methods https://doi.org/10.1038/s41592-025-02890-1 (2025).
Fraley, S. I. et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12, 598–604 (2010).
Wu, P. H., Giri, A. & Wirtz, D. Statistical analysis of cell migration in 3D using the anisotropic persistent random walk model. Nat. Protoc. 10, 517–527 (2015).
Giri, A. et al. The Arp2/3 complex mediates multigeneration dendritic protrusions for efficient 3-dimensional cancer cell migration. FASEB J. https://doi.org/10.1096/fj.12-224352 (2013).
Jayatilaka, H. et al. EB1 and cytoplasmic dynein mediate protrusion dynamics for efficient 3-dimensional cell migration. FASEB J. https://doi.org/10.1096/fj.201700444RR (2018).
Khatau, S. B. et al. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2, 488 (2012).
Wu, P. H., Giri, A., Sun, S. X. & Wirtz, D. Three-dimensional cell migration does not follow a random walk. Proc. Natl Acad. Sci. USA 111, 3949–3954 (2014).
Stokes, C. L. & Lauffenburger, D. A. Analysis of the roles of microvessel endothelial cell random motility and chemotaxis in angiogenesis. J. Theor. Biol. 152, 377–403 (1991).
Stokes, C. L., Lauffenburger, D. A. & Williams, S. K. Migration of individual microvessel endothelial cells: Stochastic model and parameter measurement. J. Cell Sci. 99, 419–430 (1991).
Chen, H. C. Boyden chamber assay. Methods Mol. Biol. 294, 15–22 (2005).
Justus, C. R., Marie, M. A., Sanderlin, E. J. & Yang, L. V. Transwell in vitro cell migration and invasion assays. Methods Mol. Biol. 2644, 349–359 (2023).
Prolo, L. M. et al. Targeted genomic CRISPR-Cas9 screen identifies MAP4K4 as essential for glioblastoma invasion. Sci. Rep. 9, 14020 (2019).
Kendirli, A. et al. A genome-wide in vivo CRISPR screen identifies essential regulators of T cell migration to the CNS in a multiple sclerosis model. Nat. Neurosci. 26, 1713–1725 (2023).
Ammann, K. R., DeCook, K. J., Li, M. & Slepian, M. J. Migration versus proliferation as contributor to in vitro wound healing of vascular endothelial and smooth muscle cells. Exp. Cell. Res. 376, 58–66 (2019).
Grada, A., Otero-Vinas, M., Prieto-Castrillo, F., Obagi, Z. & Falanga, V. Research techniques made simple: analysis of collective cell migration using the wound healing assay. J. Invest. Dermatol. 137, e11–e16 (2017).
Keese, C. R., Wegener, J., Walker, S. R. & Giaever, I. Electrical wound-healing assay for cells in vitro. Proc. Natl Acad. Sci. USA 101, 1554–1559 (2004).
Riahi, R., Yang, Y., Zhang, D. D. & Wong, P. K. Advances in wound-healing assays for probing collective cell migration. J. Lab Autom. 17, 59–65 (2012).
Maška, M. et al. The Cell Tracking Challenge: 10 years of objective benchmarking. Nat. Methods 20, 1010–1020 (2023).
Ulman, V. et al. An objective comparison of cell-tracking algorithms. Nat. Methods 14, 1141–1152 (2017).
Tinevez, J. Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Uka, A., Tare, A., Polisi, X. & Panci, I. FASTER R-CNN for cell counting in low contrast microscopic images. in Proceedings of the 2020 International Conference on Computing, Networking, Telecommunications and Engineering Sciences Applications, CoNTESA 2020 64–69 https://doi.org/10.1109/CONTESA50436.2020.9302852 (2020).
Buggenthin, F. et al. An automatic method for robust and fast cell detection in bright field images from high-throughput microscopy. BMC Bioinformatics 14, 297 (2013).
Ershov, D. et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022).
Shim, C. et al. CellTrackVis: interactive browser-based visualization for analyzing cell trajectories and lineages. BMC Bioinformatics 24, 124 (2023).
Collins, T. J. ImageJ for microscopy. Biotechniques 43, 25–30 (2007).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Maity, D. et al. Profiling dynamic patterns of single-cell motility. Adv. Sci. 11, 2400918 (2024).
Emami, N., Sedaei, Z. & Ferdousi, R. Computerized cell tracking: current methods, tools and challenges. Vis. Informatics 5, 1–13 (2021).
Wu, P. -H. et al. Single-cell morphology encodes metastatic potential. Sci. Adv. 6, eaaw6938 (2020).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol https://doi.org/10.1371/journal.pbio.2005970 (2018).
Magidson, V. & Khodjakov, A. Circumventing photodamage in live-cell microscopy. Methods Cell. Biol. 114, 545–560 (2013).
Laissue, P. P., Alghamdi, R. A., Tomancak, P., Reynaud, E. G. & Shroff, H. Assessing phototoxicity in live fluorescence imaging. Nat. Methods 14, 657–661 (2017).
Icha, J., Weber, M., Waters, J. C. & Norden, C. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays 39, 1700003 (2017).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2020).
Copied TextX Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science Vol. 11071, 265–273 (Springer, 2018).
Han, K. S. et al. qMAP enabled microanatomical mapping of human skin aging. Preprint at bioRxiv https://doi.org/10.1101/2024.04.03.588011 (2024).
Cheezum, M. K., Walker, W. F. & Guilford, W. H. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 81, 2378–2388 (2001).
Wen, C. et al. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. Elife 10, e59187 (2021).
Miller, M. J., Wei, S. H., Cahalan, M. D. & Parker, I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc. Natl Acad. Sci. USA 100, 2604–2609 (2003).
Lau, D. et al. Intravital imaging of adoptive T-cell morphology, mobility and trafficking following immune checkpoint inhibition in a mouse melanoma model. Front. Immunol. 11, 538523 (2020).
Wiggins, L. et al. The CellPhe toolkit for cell phenotyping using time-lapse imaging and pattern recognition. Nat. Commun. 14, 1854 (2023).
Shannon, M. J., Eisman, S. E., Lowe, A. R., Sloan, T. F. W. & Mace, E. M. cellPLATO – an unsupervised method for identifying cell behaviour in heterogeneous cell trajectory data. J. Cell Sci. 137, jcs261887 (2024).
Freckmann, E. C. et al. Traject3D allows label-free identification of distinct co-occurring phenotypes within 3D culture by live imaging. Nat. Commun. 13, 5317 (2022).
GitHub. quantixed/TrackMateR: analysis of TrackMate XML outputs in R. https://github.com/quantixed/TrackMateR
Wortel, I. M. N. et al. CelltrackR: An R package for fast and flexible analysis of immune cell migration data. Immunoinformatics 1–2, 100003 (2021).
Gómez-De-Mariscal, E. et al. CellTracksColab is a platform that enables compilation, analysis, and exploration of cell tracking data. PLoS Biol. 22, e3002740 (2024).
Masuzzo, P., Van Troys, M., Ampe, C. & Martens, L. Taking aim at moving targets in computational cell migration. Trends Cell Biol. 26, 88–110 (2016).
Meijering, E., Dzyubachyk, O. & Smal, I. Methods for cell and particle tracking. Methods Enzymol. 504, 183–200 (2012).
Wu, P. H. et al. Particle tracking microrheology of cancer cells in living subjects. Mater. Today 39, 98–109 (2020).
Savin, T. & Doyle, P. S. Static and dynamic errors in particle tracking microrheology. Biophys. J. 88, 623–638 (2005).
Wu, P. -H., Arce, S. H., Burney, P. R. & Tseng, Y. A novel approach to high accuracy of video-based microrheology. Biophys. J. 96, 5103–5111 (2009).
Metzner, C. et al. Superstatistical analysis and modelling of heterogeneous random walks. Nat. Commun. 6, 7516 (2015).
Lan, T. et al. Decomposition of cell activities revealing the role of the cell cycle in driving biofunctional heterogeneity. Sci. Rep. 11, 23431 (2021).
Phillip, J. M. et al. Fractional re-distribution among cell motility states during ageing. Commun. Biol. 4, 81 (2021).
Lauffenburger, D. A. & Horwitz, A. F. Cell migration: a physically integrated molecular process. Cell 84, 359–369 (1996).
Pollard, T. D. & Borisy, G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 (2003).
Ridley, A. J. et al. Cell migration: integrating signals from front to back. Science 302, 1704–1709 (2003).
Lee, M. H. et al. Mismatch in mechanical and adhesive properties induces pulsating cancer cell migration in epithelial monolayer. Biophys. J. 102, 2731–2741 (2012).
Trepat, X. et al. Physical forces during collective cell migration. Nat. Phys. 5, 426–430 (2009).
Tambe, D. T. et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10, 469–475 (2011).
Dieterich, P., Klages, R., Preuss, R. & Schwab, A. Anomalous dynamics of cell migration. Proc. Natl Acad. Sci. USA 105, 459–463 (2008).
Tranquillo, R. T., Lauffenburger, D. A. & Zigmond, S. H. A stochastic model for leukocyte random motility and chemotaxis based on receptor binding fluctuations. J. Cell Biol. 106, 303–309 (1988).
Wu, P. -H., Gilkes, D. M. & Wirtz, D. The biophysics of 3D cell migration. Annu. Rev. Biophys. 47, 549–567 (2018).
Takagi, H., Sato, M. J., Yanagida, T. & Ueda, M. Functional analysis of spontaneous cell movement under different physiological conditions. PLoS ONE 3, e2648 (2008).
Selmeczi, D., Mosler, S., Hagedorn, P. H., Larsen, N. B. & Flyvbjerg, H. Cell motility as persistent random motion: theories from experiments. Biophys. J. 89, 912–931 (2005).
Czirok, A., Schlett, K., Madarasz, E. & Vicsek, T. Exponential distribution of locomotion activity in cell cultures. Phys. Rev. Lett. 81, 3038–3041 (1999).
Campos, D., Méndez, V. & Llopis, I. Persistent random motion: uncovering cell migration dynamics. J. Theor. Biol. 267, 526–534 (2010).
Harris, T. H. et al. Generalized Lévy walks and the role of chemokines in migration of effector CD8+ T cells. Nature 486, 545–548 (2012).
Huda, S. et al. Lévy-like movement patterns of metastatic cancer cells revealed in microfabricated systems and implicated in vivo. Nat. Commun. 9, 4539 (2018).
Mantegna, R. N. & Stanley, H. E. Stochastic process with ultraslow convergence to a Gaussian: the truncated Lévy flight. Phys. Rev. Lett. 73, 2946 (1994).
Godet, I. et al. Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis. Nat. Commun. 10, 4862 (2019).
Nair, P. R. et al. MLL1 regulates cytokine-driven cell migration and metastasis. Sci. Adv. 10, eadk0785 (2024).
Du, W. et al. High-motility pro-tumorigenic monocytes drive macrophage enrichment in the tumor microenvironment. Preprint at bioRxiv https://doi.org/10.1101/2024.07.16.603739 (2024).
SenGupta, S., Parent, C. A. & Bear, J. E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 22, 529–547 (2021).
Kimmel, J. C., Chang, A. Y., Brack, A. S. & Marshall, W. F. Inferring cell state by quantitative motility analysis reveals a dynamic state system and broken detailed balance. PLoS Comput. Biol. 14, e1005927 (2018).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Ascione, F. et al. Comparison between fibroblast wound healing and cell random migration assays in vitro. Exp. Cell. Res. 347, 123–132 (2016).
Tremel, A. et al. Cell migration and proliferation during monolayer formation and wound healing. Chem. Eng. Sci. 64, 247–253 (2009).
Zordan, M. D., Mill, C. P., Riese, D. J. & Leary, J. F. A high throughput, interactive imaging, bright-field wound healing assay. Cytometry A 79, 227–232 (2011).
Jonkman, J. E. N. et al. An introduction to the wound healing assay using live-cell microscopy. Cell Adh. Migr. 8, 440–451 (2014).
Kauanova, S., Urazbayev, A. & Vorobjev, I. The frequent sampling of wound scratch assay reveals the ‘opportunity’ window for quantitative evaluation of cell motility-impeding drugs. Front. Cell Dev. Biol. 9, 640972 (2021).
Main, K. A., Mikelis, C. M. & Doçi, C. L. In vitro wound healing assays to investigate epidermal migration. Methods Mol. Biol. 2109, 147–154 (2020).
Chung, H. H., Bellefeuille, S. D., Miller, H. N. & Gaborski, T. R. Extended live-tracking and quantitative characterization of wound healing and cell migration with SiR-Hoechst. Exp. Cell. Res. 373, 198–210 (2018).
Bise, R., Kanade, T., Yin, Z. & Huh, S. Il. Automatic cell tracking applied to analysis of cell migration in wound healing assay. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2011, 6174–6179 (2011).
Poujade, M. et al. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl Acad. Sci. USA 104, 15988–15993 (2007).
Hulkower, K. I. & Herber, R. L. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 3, 107–124 (2011).
Pijuan, J. et al. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol. 7, 449183 (2019).
Justus, C. R., Leffler, N., Ruiz-Echevarria, M. & Yang, L. V. In vitro cell migration and invasion assays. J. Vis. Exp. https://doi.org/10.3791/51046 (2014).
O’brien, J., Hayder, H. & Peng, C. Automated quantification and analysis of cell counting procedures using ImageJ plugins. J. Vis. Exp. 2016, e54719 (2016).
Bird, C. & Kirstein, S. Real-time, label-free monitoring of cellular invasion and migration with the xCELLigence system. Nat. Methods https://doi.org/10.1038/nmeth.f.263 (2009).
Vinci, M. et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 10, 29 (2012).
Russo, G. C. et al. E-cadherin interacts with EGFR resulting in hyper-activation of ERK in multiple models of breast cancer. Oncogene 43, 1445–1462 (2024).
Lee, M. H. et al. Multi-compartment tumor organoids. Mater. Today 61, 104–116 (2022).
Puls, T. J. et al. Development of a novel 3D tumor-tissue invasion model for high-throughput, high-content phenotypic drug screening. Sci. Rep. 8, 13039 (2018).
Randriamanantsoa, S. et al. Spatiotemporal dynamics of self-organized branching in pancreas-derived organoids. Nat. Commun. 13, 5219 (2022).
Goranci-Buzhala, G. et al. Rapid and efficient invasion assay of glioblastoma in human brain organoids. Cell Rep. 31, 107738 (2020).
Friedrich, J., Seidel, C., Ebner, R. & Kunz-Schughart, L. A. Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 4, 309–324 (2009).
Vinci, M., Box, C. & Eccles, S. A. Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp. 2015, 52686 (2015).
Chen, Z. et al. Automated evaluation of tumor spheroid behavior in 3D culture using deep learning-based recognition. Biomaterials 272, 120770 (2021).
Heiss, J. & Tavana, H. Automated analysis of extracellular matrix invasion of cancer cells from tumor spheroids. ACS Meas. Sci. Au 4, 260–266 (2024).
Konen, J. et al. Image-guided genomics of phenotypically heterogeneous populations reveals vascular signalling during symbiotic collective cancer invasion. Nat. Commun. 8, 15078 (2017).
Valencia, A. M. J. et al. Collective cancer cell invasion induced by coordinated contractile stresses. Oncotarget 6, 43438–43451 (2015).
Kim, B. J. & Wu, M. Microfluidics for mammalian cell chemotaxis. Ann. Biomed. Eng. 40, 1316–1327 (2012).
Chen, Y. C. et al. Single-cell migration chip for chemotaxis-based microfluidic selection of heterogeneous cell populations. Sci. Rep. 5, 9980 (2015).
Yankaskas, C. L. et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 3, 452–465 (2019).
Stroka, K. M. et al. Water permeation drives tumor cell migration in confined microenvironments. Cell 157, 611–623 (2014).
Jiang, X., Bruzewicz, D. A., Wong, A. P., Piel, M. & Whitesides, G. M. Directing cell migration with asymmetric micropatterns. Proc. Natl Acad. Sci. USA 102, 975–978 (2005).
Bhattacharya, S. et al. A high-throughput microfabricated platform for rapid quantification of metastatic potential. Sci. Adv. 10, eadk0015 (2024).
Wu, J., Wu, X. & Lin, F. Recent developments in microfluidics-based chemotaxis studies. Lab Chip 13, 2484–2499 (2013).
Sackmann, E. K., Fulton, A. L. & Beebe, D. J. The present and future role of microfluidics in biomedical research. Nature 507, 181–189 (2014).
Li, J. & Lin, F. Microfluidic devices for studying chemotaxis and electrotaxis. Trends Cell Biol. 21, 489–497 (2011).
Sun, Y. S. Studying electrotaxis in microfluidic devices. Sensors 17, 2048 (2017).
Li, Y. et al. Cell migration microfluidics for electrotaxis-based heterogeneity study of lung cancer cells. Biosens. Bioelectron. 89, 837–845 (2017).
Sunyer, R. & Trepat, X. Durotaxis. Curr. Biol. 30, R383–R387 (2020).
Style, R. W. et al. Patterning droplets with durotaxis. Proc. Natl Acad. Sci. USA 110, 12541–12544 (2013).
Fanfone, D. et al. Confined migration promotes cancer metastasis through resistance to anoikis and increased invasiveness. Elife 11, e73150 (2022).
Paul, C. D., Mistriotis, P. & Konstantopoulos, K. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer 17, 131–140 (2016).
Zengel, P. et al. μ-slide chemotaxis: a new chamber for long-term chemotaxis studies. BMC Cell Biol. 12, 21 (2011).
Lambert, B. S. et al. A microfluidics-based in situ chemotaxis assay to study the behaviour of aquatic microbial communities. Nat. Microbiol. 2, 1344–1349 (2017).
Sacan, A., Ferhatosmanoglu, H. & Coskun, H. CellTrack: an open-source software for cell tracking and motility analysis. Bioinformatics 24, 1647–1649 (2008).
Hu, T., Xu, S., Wei, L., Zhang, X. & Wang, X. CellTracker: an automated toolbox for single-cell segmentation and tracking of time-lapse microscopy images. Bioinformatics 37, 285–287 (2021).
Kamentsky, L. et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics 22, 433 (2021).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Aragaki, H., Ogoh, K., Kondo, Y. & Aoki, K. LIM Tracker: a software package for cell tracking and analysis with advanced interactivity. Sci. Rep. 12, 2702 (2022).
Lugagne, J. -B., Lin, H. & Dunlop, M. J. DeLTA: automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 16, e1007673 (2020).
Zhao, Y. et al. HFM-Tracker: a cell tracking algorithm based on hybrid feature matching. Analyst 149, 2629–2636 (2024).
Deforet, M. et al. Automated velocity mapping of migrating cell populations (AVeMap). Nat. Methods 9, 1081–1083 (2012).
Amat, F. et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951–958 (2014).
Cordelières, F. P. et al. Automated cell tracking and analysis in phase-contrast videos (iTrack4U): development of Java software based on combined mean-shift processes. PLoS ONE 8, e81266 (2013).
Masuzzo, P. et al. CellMissy: a tool for management, storage and analysis of cell migration data produced in wound healing-like assays. Bioinformatics 29, 2661–2663 (2013).
Gebäck, T., Schulz, M. M. P., Koumoutsakos, P. & Detmar, M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 46, 265–274 (2009).
Garcia-Fossa, F., Gaal, V. & de Jesus, M. B. PyScratch: an ease of use tool for analysis of scratch assays. Comput. Methods Programs Biomed. 193, 105476 (2020).
Cortesi, M., Pasini, A., Tesei, A. & Giordano, E. AIM: a computational tool for the automatic quantification of scratch wound healing assays. Appl. Sci. 7, 1237 (2017).
Li, X., Yang, H., Huang, H. & Zhu, T. CELLCOUNTER: novel open-source software for counting cell migration and invasion in vitro. Biomed. Res. Int. 2014, 863564 (2014).
Cortesi, M. et al. I-AbACUS: a reliable software tool for the semi-automatic analysis of invasion and migration Transwell assays. Sci. Rep. 8, 3814 (2018).
Chen, W. et al. High-throughput image analysis of tumor spheroids: a user-friendly software application to measure the size of spheroids automatically and accurately. J. Vis. Exp. https://doi.org/10.3791/51639 (2014).
Piccinini, F. AnaSP: a software suite for automatic image analysis of multicellular spheroids. Comput. Methods Prog. Biomed. 119, 43–52 (2015).
Akshay, A. et al. SpheroScan: a user-friendly deep learning tool for spheroid image analysis. Gigascience 12, giad082 (2022).
Park, T. et al. Development of a deep learning based image processing tool for enhanced organoid analysis. Sci. Rep. 13, 19841 (2023).
Matthews, J. M. et al. OrganoID: a versatile deep learning platform for tracking and analysis of single-organoid dynamics. PLoS Comput. Biol. 18, e1010584 (2022).
Acknowledgements
We acknowledge the following sources of support: U54AR081774 (to D.W.), U54CA268083 (to D.W.), R01CA300052 (to D.W.), UG3CA275681 (to P.-H.W.) and UH3CA275681 (to P.-H.W.), all from the National Cancer Institute; R35-GM157099 (to J.M.P.) from the National Institute of General Medical Sciences; and an American Federation for Aging Research Glenn Foundation Junior Faculty Award (to J.M.P.).
Author information
Authors and Affiliations
Contributions
D.W., P.-H.W. and J.M.P. conceived the outline. P.-H.W., J.M.P., W.D., A.F., P.R.N. and D.W. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Li Yang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Table 1
Mathematical definitions and common metrics used to analyze cell trajectories.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, PH., Phillip, J.M., Du, W. et al. Methods to analyze cell migration data: fundamentals and practical guidelines. Nat Methods 23, 43–55 (2026). https://doi.org/10.1038/s41592-025-02935-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02935-5
This article is cited by
-
Selecting the optimal cell migration assay: fundamentals and practical guidelines
Nature Methods (2026)