Abstract
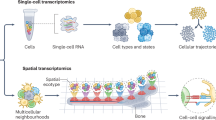
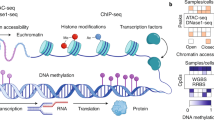
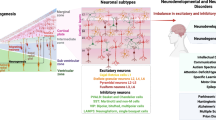
Over the past decade, single-cell genomics technologies have allowed scalable profiling of cell-type-specific features, which has substantially increased our ability to study cellular diversity and transcriptional programs in heterogeneous tissues. Yet our understanding of mechanisms of gene regulation or the rules that govern interactions between cell types is still limited. The advent of new computational pipelines and technologies, such as single-cell epigenomics and spatially resolved transcriptomics, has created opportunities to explore two new axes of biological variation: cell-intrinsic regulation of cell states and expression programs and interactions between cells. Here, we summarize the most promising and robust technologies in these areas, discuss their strengths and limitations and discuss key computational approaches for analysis of these complex datasets. We highlight how data sharing and integration, documentation, visualization and benchmarking of results contribute to transparency, reproducibility, collaboration and democratization in neuroscience, and discuss needs and opportunities for future technology development and analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
16 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41593-024-01858-2
References
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
Kim, S. S. et al. Leveraging single-cell ATAC-seq and RNA-seq to identify disease-critical fetal and adult brain cell types. Nat. Commun. 15, 563 (2024).
Sun, N. et al. Single-nucleus multiregion transcriptomic analysis of brain vasculature in Alzheimer’s disease. Nat. Neurosci. 26, 970–982 (2023).
Cain, A. et al. Multicellular communities are perturbed in the aging human brain and Alzheimer’s disease. Nat. Neurosci. 26, 1267–1280 (2023).
Kim, C. N., Shin, D., Wang, A. & Nowakowski, T. J. Spatiotemporal molecular dynamics of the developing human thalamus. Science 382, eadf9941 (2023).
Pineda, S. S. et al. Single-cell dissection of the human motor and prefrontal cortices in ALS and FTLD. Cell 187, 1971–1989 (2024).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Green, G. S. et al. Cellular communities reveal trajectories of brain ageing and Alzheimer’s disease. Nature https://doi.org/10.1038/s41586-024-07871-6 (2024).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Davis, A., Gao, R. & Navin, N. E. SCOPIT: sample size calculations for single-cell sequencing experiments. BMC Bioinformatics 20, 566 (2019).
Schmid, K. T. et al. scPower accelerates and optimizes the design of multi-sample single cell transcriptomic studies. Nat. Commun. 12, 6625 (2021).
Su, K., Wu, Z. & Wu, H. Simulation, power evaluation and sample size recommendation for single-cell RNA-seq. Bioinformatics 36, 4860–4868 (2020).
Phipson, B. et al. Propeller: testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726 (2022).
Lin, Y. et al. scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Mol. Syst. Biol. 16, e9389 (2020).
Jeon, H. et al. Statistical power analysis for designing bulk, single-cell, and spatial transcriptomics experiments: review, tutorial, and perspectives. Biomolecules 13, 221 (2023).
Ryaboshapkina, M. & Azzu, V. Sample size calculation for a NanoString GeoMx spatial transcriptomics experiment to study predictors of fibrosis progression in non-alcoholic fatty liver disease. Sci. Rep. 13, 8943 (2023).
Colonna, M. et al. Implementation and validation of single-cell genomics experiments in neuroscience. Nat. Neurosci. https://doi.org/10.1038/s41593-024-01814-0 (2024).
Zhang, Y. et al. Deconvolution algorithms for inference of the cell-type composition of the spatial transcriptome. Comput. Struct. Biotechnol. J. 21, 176–184 (2023).
Im, Y. & Kim, Y. A comprehensive overview of RNA deconvolution methods and their application. Mol. Cells 46, 99–105 (2023).
Charytonowicz, D., Brody, R. & Sebra, R. Interpretable and context-free deconvolution of multi-scale whole transcriptomic data with UniCell deconvolve. Nat. Commun. 14, 1350 (2023).
Chen, Y. et al. Deep autoencoder for interpretable tissue-adaptive deconvolution and cell-type-specific gene analysis. Nat. Commun. 13, 6735 (2022).
Liao, J. et al. De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution. Nat. Commun. 13, 6498 (2022).
Heimberg, G., Bhatnagar, R., El-Samad, H. & Thomson, M. Low dimensionality in gene expression data enables the accurate extraction of transcriptional programs from shallow sequencing. Cell Syst. 2, 239–250 (2016).
Haque, A., Engel, J., Teichmann, S. A. & Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75 (2017).
Boyce, R. W., Dorph-Petersen, K. -A., Lyck, L. & Gundersen, H. J. G. Design-based stereology: introduction to basic concepts and practical approaches for estimation of cell number. Toxicol. Pathol. 38, 1011–1025 (2010).
Adameyko, I. et al. Applying single-cell/nucleus genomics to studies of cellular heterogeneity and cell fate transitions in the nervous system. Nat. Neurosci. https://doi.org/10.1038/s41593-024-01827-9 (2024).
Yu, L., Cao, Y., Yang, J. Y. H. & Yang, P. Benchmarking clustering algorithms on estimating the number of cell types from single-cell RNA-sequencing data. Genome Biol. 23, 49 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Swapna, L. S., Huang, M. & Li, Y. GTM-decon: guided-topic modeling of single-cell transcriptomes enables sub-cell-type and disease-subtype deconvolution of bulk transcriptomes. Genome Biol. 24, 190 (2023).
Zhang, S., Yang, L., Yang, J., Lin, Z. & Ng, M. K. Dimensionality reduction for single cell RNA sequencing data using constrained robust non-negative matrix factorization. NAR Genom. Bioinform. 2, lqaa064 (2020).
Morabito, S., Reese, F., Rahimzadeh, N., Miyoshi, E. & Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 3, 100498 (2023).
Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Cortal, A., Martignetti, L., Six, E. & Rausell, A. Gene signature extraction and cell identity recognition at the single-cell level with Cell-ID. Nat. Biotechnol. 39, 1095–1102 (2021).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Bayraktar, O. A. et al. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509 (2020).
Shi, H. et al. Spatial atlas of the mouse central nervous system at molecular resolution. Nature 622, 552–561 (2023).
Zhang, M. et al. Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature 624, 343–354 (2023).
Langlieb, J. et al. The molecular cytoarchitecture of the adult mouse brain. Nature 624, 333–342 (2023).
Stanley, G., Gokce, O., Malenka, R. C., Südhof, T. C. & Quake, S. R. Continuous and discrete neuron types of the adult murine striatum. Neuron 105, 688–699 (2020).
Muñoz-Manchado, A. B. et al. Diversity of interneurons in the dorsal striatum revealed by single-cell RNA sequencing and PatchSeq. Cell Rep. 24, 2179–2190 (2018).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022).
Ghazanfar, S., Guibentif, C. & Marioni, J. C. Stabilized mosaic single-cell data integration using unshared features. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01766-z (2023).
Mages, S. et al. TACCO unifies annotation transfer and decomposition of cell identities for single-cell and spatial omics. Nat. Biotechnol. 41, 1465–1473 (2023).
Lohoff, T. et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat. Biotechnol. 40, 74–85 (2022).
Li, B. et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods 19, 662–670 (2022).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Baldoni, P. L. et al. Dividing out quantification uncertainty allows efficient assessment of differential transcript expression with edgeR. Nucleic Acids Res. 52, e13 (2024).
Squair, J. W. et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 12, 5692 (2021).
Hoffman, G. E. et al. Efficient differential expression analysis of large-scale single cell transcriptomics data using dreamlet. Preprint at bioRxiv https://doi.org/10.1101/2023.03.17.533005 (2023).
Hoffman, G. E. & Schadt, E. E. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483 (2016).
Gabitto, M. I. et al. Integrated multimodal cell atlas of Alzheimer’s disease. Nat. Neurosci. https://doi.org/10.1038/s41593-024-01774-5 (2024).
Zeng, H. et al. Integrative in situ mapping of single-cell transcriptional states and tissue histopathology in a mouse model of Alzheimer’s disease. Nat. Neurosci. 26, 430–446 (2023).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 185, 1665–1681 (2020).
Ke, R. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013).
Russell, A. J. C. et al. Slide-tags enables single-nucleus barcoding for multimodal spatial genomics. Nature 625, 101–109 (2024).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Chen, X. et al. High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell 179, 772–786 (2019).
Condylis, C. et al. Dense functional and molecular readout of a circuit hub in sensory cortex. Science 375, eabl5981 (2022).
Li, Q. et al. Multimodal charting of molecular and functional cell states via in situ electro-sequencing. Cell 186, 2002–2017 (2023).
Deng, Y. et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Lu, T., Ang, C. E. & Zhuang, X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 185, 4448–4464 (2022).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Llorens-Bobadilla, E. et al. Solid-phase capture and profiling of open chromatin by spatial ATAC. Nat. Biotechnol. 41, 1085–1088 (2023).
Zeng, H. et al. Spatially resolved single-cell translatomics at molecular resolution. Science 380, eadd3067 (2023).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Sans, M. et al. Integrated spatial transcriptomics and lipidomics of precursor lesions of pancreatic cancer identifies enrichment of long chain sulfatide biosynthesis as an early metabolic alteration. Preprint at bioRxiv https://doi.org/10.1101/2023.08.14.553002 (2023).
Vicari, M. et al. Spatial multimodal analysis of transcriptomes and metabolomes in tissues. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01937-y (2023).
Wang, Q. et al. The Allen Mouse Brain Common Coordinate Framework: a 3D reference atlas. Cell 181, 936–953 (2020).
Clifton, K. et al. STalign: alignment of spatial transcriptomics data using diffeomorphic metric mapping. Nat. Commun. 14, 8123 (2023).
Ortiz, C. et al. Molecular atlas of the adult mouse brain. Sci. Adv. 6, eabb3446 (2020).
Kumar, K. et al. Subcortical brain alterations in carriers of genomic copy number variants. Am. J. Psychiatry 180, 685–698 (2023).
Moreau, C. A. et al. Brain functional connectivity mirrors genetic pleiotropy in psychiatric conditions. Brain 146, 1686–1696 (2023).
Bruschi, N., Boffa, G. & Inglese, M. Ultra-high-field 7-T MRI in multiple sclerosis and other demyelinating diseases: from pathology to clinical practice. Eur. Radiol. Exp. 4, 59 (2020).
Tang, Z. et al. Search and match across spatial omics samples at single-cell resolution. Nat. Methods 21, 1818–1829 (2024).
Xia, C. -R., Cao, Z. -J., Tu, X. -M. & Gao, G. Spatial-linked alignment tool (SLAT) for aligning heterogenous slices. Nat. Commun. 14, 7236 (2023).
Hofmann, A. et al. Myeloid cell iron uptake pathways and paramagnetic rim formation in multiple sclerosis. Acta Neuropathol. 146, 707–724 (2023).
Sucksdorff, M. et al. Brain TSPO-PET predicts later disease progression independent of relapses in multiple sclerosis. Brain 143, 3318–3330 (2020).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Fang, R. et al. Conservation and divergence of cortical cell organization in human and mouse revealed by MERFISH. Science 377, 56–62 (2022).
Wu, S. J. et al. Cortical somatostatin interneuron subtypes form cell-type-specific circuits. Neuron 111, 2675–2692 (2023).
Garcia-Alonso, L. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 53, 1698–1711 (2021).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Tehranchi, A. et al. Fine-mapping cis-regulatory variants in diverse human populations. Elife 8, e39595 (2019).
Kosoy, R. et al. Genetics of the human microglia regulome refines Alzheimer’s disease risk loci. Nat. Genet. 54, 1145–1154 (2022).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Cusanovich, D. A. et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Gontarz, P. et al. Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci. Rep. 10, 10150 (2020).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Grandi, F. C., Modi, H., Kampman, L. & Corces, M. R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 17, 1518–1552 (2022).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Macneil, L. T. & Walhout, A. J. M. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 21, 645–657 (2011).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17, 147–154 (2020).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Bravo González-Blas, C. et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116 (2020).
Kartha, V. K. et al. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2, 100166 (2022).
Bravo González-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 20, 1355–1367 (2023).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Lynch, A. W. et al. MIRA: joint regulatory modeling of multimodal expression and chromatin accessibility in single cells. Nat. Methods 19, 1097–1108 (2022).
Xuan, C. et al. scBPGRN: integrating single-cell multi-omics data to construct gene regulatory networks based on BP neural network. Comput. Biol. Med. 151, 106249 (2022).
Kang, J. B. et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat. Commun. 12, 5890 (2021).
Macaulay, I. C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, e21856 (2017).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Schmid, M., Durussel, T. & Laemmli, U. K. ChIC and ChEC; genomic mapping of chromatin proteins. Mol. Cell 16, 147–157 (2004).
Grosselin, K. et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060–1066 (2019).
Wu, S. J. et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 39, 819–824 (2021).
Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216 (2019).
Handa, T. et al. Chromatin integration labeling for mapping DNA-binding proteins and modifications with low input. Nat. Protoc. 15, 3334–3360 (2020).
Bartosovic, M., Kabbe, M. & Castelo-Branco, G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 39, 825–835 (2021).
Bartosovic, M. & Castelo-Branco, G. Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag. Nat. Biotechnol. 41, 794–805 (2023).
Gopalan, S., Wang, Y., Harper, N. W., Garber, M. & Fazzio, T. G. Simultaneous profiling of multiple chromatin proteins in the same cells. Mol. Cell 81, 4736–4746 (2021).
Tedesco, M. et al. Chromatin Velocity reveals epigenetic dynamics by single-cell profiling of heterochromatin and euchromatin. Nat. Biotechnol. 40, 235–244 (2022).
Meers, M. P., Llagas, G., Janssens, D. H., Codomo, C. A. & Henikoff, S. Multifactorial profiling of epigenetic landscapes at single-cell resolution using MulTI-Tag. Nat. Biotechnol. 41, 708–716 (2023).
Li, C., Virgilio, M. C., Collins, K. L. & Welch, J. D. Multi-omic single-cell velocity models epigenome-transcriptome interactions and improves cell fate prediction. Nat. Biotechnol. 41, 387–398 (2023).
Schoenfelder, S. & Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Noack, F. et al. Multimodal profiling of the transcriptional regulatory landscape of the developing mouse cortex identifies Neurog2 as a key epigenome remodeler. Nat. Neurosci. 25, 154–167 (2022).
Zuin, J. et al. Nonlinear control of transcription through enhancer-promoter interactions. Nature 604, 571–577 (2022).
Spielmann, M., Lupiáñez, D. G. & Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467 (2018).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Tan, L., Xing, D., Chang, C. -H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Tan, L. et al. Changes in genome architecture and transcriptional dynamics progress independently of sensory experience during post-natal brain development. Cell 184, 741–758 (2021).
Zhou, T. et al. GAGE-seq concurrently profiles multiscale 3D genome organization and gene expression in single cells. Nat. Genet. https://doi.org/10.1038/s41588-024-01745-3 (2024).
Wu, H. & Zhang, Y. Charting oxidized methylcytosines at base resolution. Nat. Struct. Mol. Biol. 22, 656–661 (2015).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013).
Luo, C., Hajkova, P. & Ecker, J. R. Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340 (2018).
Luo, C. et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017).
Iqbal, W. & Zhou, W. Computational methods for single-cell DNA methylome analysis. Genomics Proteomics Bioinformatics 21, 48–66 (2023).
Smallwood, S. A. et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014).
Luo, C. et al. Robust single-cell DNA methylome profiling with snmC-seq2. Nat. Commun. 9, 3824 (2018).
Lee, D. -S. et al. Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nat. Methods 16, 999–1006 (2019).
Luo, C. et al. Single nucleus multi-omics identifies human cortical cell regulatory genome diversity. Cell Genom. 2, 100107 (2022).
Fabyanic, E. B. et al. Quantitative single cell 5hmC sequencing reveals non-canonical gene regulation by non-CG hydroxymethylation. Preprint at bioRxiv https://doi.org/10.1101/2021.03.23.434325 (2021).
Nichols, R. V. et al. High-throughput robust single-cell DNA methylation profiling with sciMETv2. Nat. Commun. 13, 7627 (2022).
Abdulhay, N. J. et al. Massively multiplex single-molecule oligonucleosome footprinting. Elife 9, e59404 (2020).
Stergachis, A. B., Debo, B. M., Haugen, E., Churchman, L. S. & Stamatoyannopoulos, J. A. Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science 368, 1449–1454 (2020).
Shipony, Z. et al. Long-range single-molecule mapping of chromatin accessibility in eukaryotes. Nat. Methods 17, 319–327 (2020).
Lee, I. et al. Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. Nat. Methods 17, 1191–1199 (2020).
Wang, Y. et al. Single-molecule long-read sequencing reveals the chromatin basis of gene expression. Genome Res. 29, 1329–1342 (2019).
Sharon, D., Tilgner, H., Grubert, F. & Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31, 1009–1014 (2013).
Gupta, I. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 36, 1197–1202 (2018).
Isaac, R. S. et al. Single-nucleoid architecture reveals heterogeneous packaging of mitochondrial DNA. Nat. Struct. Mol. Biol. 31, 568–577 (2024).
Abdulhay, N. J. et al. Nucleosome density shapes kilobase-scale regulation by a mammalian chromatin remodeler. Nat. Struct. Mol. Biol. 30, 1571–1581 (2023).
Marconato, L. et al. SpatialData: an open and universal data framework for spatial omics. Nat. Methods https://doi.org/10.1038/s41592-024-02212-x (2024).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5, e12776 (2010).
Kim, S. Ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22, 665–674 (2015).
Specht, A. T. & Li, J. LEAP: constructing gene co-expression networks for single-cell RNA-sequencing data using pseudotime ordering. Bioinformatics 33, 764–766 (2017).
Moerman, T. et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics 35, 2159–2161 (2019).
Deshpande, A., Chu, L. -F., Stewart, R. & Gitter, A. Network inference with Granger causality ensembles on single-cell transcriptomics. Cell Rep. 38, 110333 (2022).
Margolin, A. A. et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7, S7 (2006).
Chan, T. E., Stumpf, M. P. H. & Babtie, A. C. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 5, 251–267 (2017).
Qiu, X. et al. Inferring causal gene regulatory networks from coupled single-cell expression dynamics using Scribe. Cell Syst. 10, 265–274 (2020).
Faith, J. J. et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5, e8 (2007).
Sanchez-Castillo, M., Blanco, D., Tienda-Luna, I. M., Carrion, M. C. & Huang, Y. A Bayesian framework for the inference of gene regulatory networks from time and pseudo-time series data. Bioinformatics 34, 964–970 (2018).
Yu, J., Smith, V. A., Wang, P. P., Hartemink, A. J. & Jarvis, E. D. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics 20, 3594–3603 (2004).
Dojer, N., Bednarz, P., Podsiadlo, A. & Wilczynski, B. BNFinder2: faster Bayesian network learning and Bayesian classification. Bioinformatics 29, 2068–2070 (2013).
Wilczyński, B. & Dojer, N. BNFinder: exact and efficient method for learning Bayesian networks. Bioinformatics 25, 286–287 (2009).
Woodhouse, S., Piterman, N., Wintersteiger, C. M., Göttgens, B. & Fisher, J. SCNS: a graphical tool for reconstructing executable regulatory networks from single-cell genomic data. BMC Syst. Biol. 12, 59 (2018).
Yuan, Y. & Bar-Joseph, Z. Deep learning of gene relationships from single cell time-course expression data. Brief. Bioinform. 22, bbab142 (2021).
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
Polychronidou, M. et al. Single‐cell biology: what does the future hold? Mol. Syst. Biol. 19, e11799 (2023).
Matsumoto, H. et al. SCODE: an efficient regulatory network inference algorithm from single-cell RNA-seq during differentiation. Bioinformatics 33, 2314–2321 (2017).
Aubin-Frankowski, P. -C. & Vert, J. -P. Gene regulation inference from single-cell RNA-seq data with linear differential equations and velocity inference. Bioinformatics 36, 4774–4780 (2020).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in Single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Bais, A. S. & Kostka, D. scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 36, 1150–1158 (2020).
Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9, giaa151 (2020).
Mohammad, N. S., Nazli, R., Zafar, H. & Fatima, S. Effects of lipid based Multiple Micronutrients Supplement on the birth outcome of underweight pre-eclamptic women: a randomized clinical trial. Pak. J. Med. Sci. Q. 38, 219–226 (2022).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Qian, X. et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat. Methods 17, 101–106 (2020).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Janesick, A. et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue. Nat. Commun. https://doi.org/10.1038/s41467-023-43458-x (2023).
Kukanja, P. et al. Cellular architecture of evolving neuroinflammatory lesions and multiple sclerosis pathology. Cell 187, 1990–2009 (2024).
Missarova, A. et al. geneBasis: an iterative approach for unsupervised selection of targeted gene panels from scRNA-seq. Genome Biol. 22, 333 (2021).
Nelson, M. E., Riva, S. G. & Cvejic, A. SMaSH: a scalable, general marker gene identification framework for single-cell RNA-sequencing. BMC Bioinformatics 23, 328 (2022).
Borm, L. E. et al. Scalable in situ single-cell profiling by electrophoretic capture of mRNA using EEL FISH. Nat. Biotechnol. 41, 222–231 (2023).
Fischer, D. S., Schaar, A. C. & Theis, F. J. Modeling intercellular communication in tissues using spatial graphs of cells. Nat. Biotechnol. 41, 332–336 (2023).
Acknowledgements
We thank I. Adameyko, R. Awatramani, T. Bakken, H. S. Bateup, A. Bhaduri, C. R. Cadwell, E. Caglayan, J. L. Chen, C. Chhatbar, M. G. Filbin, D. Gate, J. Gillis, H. Hochgerner, M. Kampmann, C. N. Kim, F. Krienen, J. Krull, G. La Manno, S. Marsh, M. Monje, Q. Li, Sten Linnarsson, Q. Ma, C. Mayer, V. Menon, P. Nano, M. R O’Dea, R. Patani, A. Pollen, M. Prinz, S. Quake, F. J. Quintana, M. Scavuzzo, M. Schmitz, S. Sloan, P. Tesar, J. Tollkuhn, M. Antonietta Tosches, M. E. Urbanek, C. Walsh, J. Werner and J. Yang for the insightful feedback on this work. G.C.-B. was supported by the Swedish Research Council (grant 2019-01360), Knut and Alice Wallenberg Foundation (grants 2019-0107 and 2019-0089), The Swedish Brain Foundation (FO2023-0032), The Swedish Society for Medical Research (SSMF, grant JUB2019) and the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine. We acknowledge National Institutes of Health (NIH) grants U01AG072573, P01AG073082 and UM1HG012076 (to M.R.C.) and U01DA052713, RF1AG079557, RF1NS128908 and R01AG079291 (to Y.S.). We also acknowledge NIH awards R01MH125516, R01NS123263, R01MH128364 and U01MH130962, the Esther A & Joseph Klingenstein Fund (to T.J.N.), the Shurl and Kay Curci Foundation (T.J.N.) and the Sontag Foundation (to T.J.N.) and a gift from the William K. Bowes Jr Foundation. T.J.N. is a New York Stem Cell Foundation Robertson Neuroscience Investigator. We acknowledge awards from the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition (220020467), the Simons Foundation (947591), NHGRI (HG011641), NINDS (NS115821, NS126143) and NIMH (MH126481, MH103517) to G.K. M.H. received a JPB Foundation Picower Institute Innovation Fund award. Work in the group of B.B. was supported by the Helmholtz Center Munich, DFG priority program SPP2202 (BO 5516/1-1), ERA-NET Neuron (MOSAIC) and European Research Council Consolidator grant to B.B. (EpiCortex, 101044469). M.N. acknowledges support from the Israel Science Foundation (grant no. 1079/21), and the European Union (ERC, DecodeSC, 101040660). O.A.B. acknowledges Wellcome Sanger core and Wellcome LEAP funding. J.F. acknowledges support from the National Institute of General Medical Sciences of the NIH under award number R35-GM142889. We acknowledge NINDS Intramural funds through 1ZIA NS003153 (to A.J.L.). K.R.M. acknowledges support from the Lieber Institute for Brain Development and NIH (R01DA055823). L.S. acknowledges support from the European Research Council (ERC StG ‘DecOmPress’, 950584), the National Multiple Sclerosis Society (PA-2022-36405 and RFA-2203-39300) and the German Research Foundation through collaborative research projects (SPP 2395, FOR 2690 and GRK 2727), individual research grants (SCHI 1330/4-1 and SCHI 1330/10-1) and a Heisenberg Fellowship (SCHI 1330/6-1). N.H. acknowledges support from the Israel Science Foundation research grant no. 1709/19 and the European Research Council grant 853409. S.A.L. acknowledges support from the NIH (R01EY033353), the Cure Alzheimer’s Fund, the Belfer Neurodegeneration Consortium, HHMI Emerging Pathogens Initiative and the Carol and Gene Ludwig Family Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
S.A.L. declares a financial interest in AstronauTx and Synapticure. All other authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Christine Cheng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bonev, B., Castelo-Branco, G., Chen, F. et al. Opportunities and challenges of single-cell and spatially resolved genomics methods for neuroscience discovery. Nat Neurosci 27, 2292–2309 (2024). https://doi.org/10.1038/s41593-024-01806-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-024-01806-0
This article is cited by
-
Research progress of single cell RNA sequencing in nervous system
Molecular Biology Reports (2026)
-
Single-cell sequencing and transcriptomic data reveal that P65 activation significantly promotes microglia-mediated neuroinflammation after ischemic stroke
Scientific Reports (2025)
-
High-parameter spatial multi-omics through histology-anchored integration
Nature Methods (2025)
-
Focus on single-cell genomics
Nature Neuroscience (2024)
-
Applying single-cell and single-nucleus genomics to studies of cellular heterogeneity and cell fate transitions in the nervous system
Nature Neuroscience (2024)