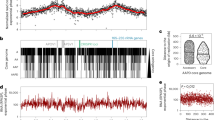
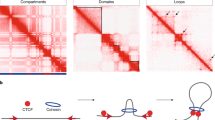
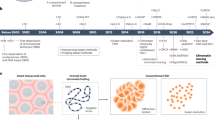
Abstract
The genome of all organisms is spatially organized to function efficiently. The advent of genome-wide chromatin conformation capture (Hi-C) methods has revolutionized our ability to probe the three-dimensional (3D) organization of genomes across diverse species. In this Review, we compare 3D chromatin folding from bacteria and archaea to that in mammals and plants, focusing on topology at the level of gene regulatory domains. In doing so, we consider systematic similarities and differences that hint at the origin and evolution of spatial chromatin folding and its relation to gene activity. We discuss the universality of spatial chromatin domains in all kingdoms, each encompassing one to several genes. We also highlight differences between organisms and suggest that similar features in Hi-C matrices do not necessarily reflect the same biological process or function. Furthermore, we discuss the evolution of domain boundaries and boundary-forming proteins, which indicates that structural maintenance of chromosome (SMC) proteins and the transcription machinery are the ancestral sculptors of the genome. Architectural proteins such as CTCF serve as clade-specific determinants of genome organization. Finally, studies in many non-model organisms show that, despite the ancient origin of 3D chromatin folding and its intricate link to gene activity, evolution tolerates substantial changes in genome organization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Misteli, T. The self-organizing genome: principles of genome architecture and function. Cell 183, 28–45 (2020).
Acemel, R. D. & Lupiáñez, D. G. Evolution of 3D chromatin organization at different scales. Curr. Opin. Genet. Dev. 78, 102019 (2023).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Hoencamp, C. et al. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372, 984–989 (2021).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Vietri Rudan, M. et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Vazquez, J., Belmont, A. S. & Sedat, J. W. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11, 1227–1239 (2001).
Hübner, M. R. & Spector, D. L. Chromatin dynamics. Annu. Rev. Biophys. 39, 471–489 (2010).
Harmston, N. et al. Topologically associating domains are ancient features that coincide with metazoan clusters of extreme noncoding conservation. Nat. Commun. 8, 441 (2017).
Kikuta, H. et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 17, 545–555 (2007).
Bolt, C. C. & Duboule, D. The regulatory landscapes of developmental genes. Development 147, dev171736 (2020).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944 (2017).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320 (2017).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Cazet, J. F. et al. New Hydra genomes reveal conserved principles of hydrozoan transcriptional regulation. Genome Res. 33, 283–298 (2023).
Schmidbaur, H. et al. Emergence of novel cephalopod gene regulation and expression through large-scale genome reorganization. Nat. Commun. 13, 2172 (2022).
Lukyanchikova, V. et al. Anopheles mosquitoes reveal new principles of 3D genome organization in insects. Nat. Commun. 13, 1960 (2022).
Marlétaz, F. et al. The little skate genome and the evolutionary emergence of wing-like fins. Nature 616, 495–503 (2023).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, 6456–6465 (2015).
Fudenberg, G., Abdennur, N., Imakaev, M., Goloborodko, A. & Mirny, L. A. Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb. Symp. Quant. Biol. 82, 45–55 (2017).
Mizuguchi, T. et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 516, 432–435 (2014).
Davidson, I. F. et al. CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nature 616, 822–827 (2023).
Davidson, I. F. et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 35, 2671–2685 (2016).
Hoencamp, C. & Rowland, B. D. Genome control by SMC complexes. Nat. Rev. Mol. Cell Biol. 24, 633–650 (2023).
Heger, P., Marin, B., Bartkuhn, M., Schierenberg, E. & Wiehe, T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl Acad. Sci. USA 109, 17507–17512 (2012).
Van Bortle, K. et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15, R82 (2014).
Kahn, T. G. et al. Topological screen identifies hundreds of Cp190- and CTCF-dependent Drosophila chromatin insulator elements. Sci. Adv. 9, eade0090 (2023).
Pollex, T. et al. Chromatin gene–gene loops support the cross-regulation of genes with related function. Mol. Cell 84, 822–838 (2023).
Umbarger, M. A. et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell 44, 252–264 (2011).
Le, T. B. K., Imakaev, M. V., Mirny, L. A. & Laub, M. T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013).
Marbouty, M. et al. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. eLife 3, e03318 (2014).
Dame, R. T., Rashid, F. M. & Grainger, D. C. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet. 21, 227–242 (2020).
Trussart, M. et al. Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae. Nat. Commun. 8, 14665 (2017).
Wang, X. et al. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–1675 (2015).
Val, M.-E. et al. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci. Adv. 2, e1501914 (2016).
Lioy, V. S. et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783 (2018).
Le, T. B. & Laub, M. T. Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO J. 35, 1582–1595 (2016).
Takemata, N. & Bell, S. D. Physical and functional compartmentalization of archaeal chromosomes. Cell 179, 165–179 (2019).
Takemata, N. & Bell, S. D. Multi-scale architecture of archaeal chromosomes. Mol. Cell 81, 473–487 (2021).
Cockram, C., Thierry, A., Gorlas, A., Lestini, R. & Koszul, R. Euryarchaeal genomes are folded into SMC-dependent loops and domains, but lack transcription-mediated compartmentalization. Mol. Cell 81, 459–472 (2021).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Hsieh, T. H. S. et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119 (2015).
Oberbeckmann, E., Quililan, K., Cramer, P. & Oudelaar, A. M. In vitro reconstitution of chromatin domains shows a role for nucleosome positioning in 3D genome organization. Nat. Genet. 56, 483–492 (2024).
Tourdot, E. & Grob, S. Three-dimensional chromatin architecture in plants — general features and novelties. Eur. J. Cell Biol. 102, 151344 (2023).
Grob, S., Schmid, M. W. & Grossniklaus, U. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 55, 678–693 (2014).
Feng, S. et al. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55, 694–707 (2014).
Dong, P., Tu, X., Liang, Z., Kang, B.-H. & Zhong, S. Plant and animal chromatin three-dimensional organization: similar structures but different functions. J. Exp. Bot. 71, 5119–5128 (2020).
Liu, C., Cheng, Y., Wang, J. & Weigel, D. Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat. Plants 3, 742–748 (2017).
Maccaferri, M. et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895 (2019).
Raymond, O. et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772–777 (2018).
Jibran, R. et al. Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Hortic. Res. 5, 8 (2018).
Hu, Y. et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748 (2019).
Dong, Q. et al. Genome-wide Hi-C analysis reveals extensive hierarchical chromatin interactions in rice. Plant J. 94, 1141–1156 (2018).
Ricci, W. A. et al. Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5, 1237–1249 (2019).
Sun, L. et al. Mapping nucleosome-resolution chromatin organization and enhancer–promoter loops in plants using Micro-C-XL. Nat. Commun. 15, 35 (2024).
Dong, P. et al. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 10, 1497–1509 (2017).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433 (2017).
Lu, J. Y. et al. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 31, 613–630 (2021).
Marinov, G. K. & Lynch, M. Diversity and divergence of dinoflagellate histone proteins. G3 6, 397–422 (2015).
Wang, M. et al. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat. Plants 4, 90–97 (2018).
Li, E. et al. Long-range interactions between proximal and distal regulatory regions in maize. Nat. Commun. 10, 2633 (2019).
Phillips-Cremins, J. E. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Fraser, J. et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 11, 852 (2015).
Zhan, Y. et al. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 27, 479–490 (2017).
Long, H. S. et al. Making sense of the linear genome, gene function and TADs. Epigenetics Chromatin 15, 4 (2022).
Rinzema, N. J. et al. Building regulatory landscapes reveals that an enhancer can recruit cohesin to create contact domains, engage CTCF sites and activate distant genes. Nat. Struct. Mol. Biol. 29, 563–574 (2022).
Kane, L. et al. Cohesin is required for long-range enhancer action at the Shh locus. Nat. Struct. Mol. Biol. 29, 891–897 (2022).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Ringel, A. R. et al. Repression and 3D-restructuring resolves regulatory conflicts in evolutionarily rearranged genomes. Cell 185, 3689–3704 (2022).
Kaushal, A. et al. Essential role of Cp190 in physical and regulatory boundary formation. Sci. Adv. 8, eabl8834 (2022).
Cavalheiro, G. R. et al. CTCF, BEAF-32, and CP190 are not required for the establishment of TADs in early Drosophila embryos but have locus-specific roles. Sci. Adv. 9, eade1085 (2023).
Hou, C., Li, L., Qin, Z. S. & Corces, V. G. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484 (2012).
Ogiyama, Y., Schuettengruber, B., Papadopoulos, G. L., Chang, J.-M. & Cavalli, G. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol. Cell 71, 73–88 (2018).
Luzhin, A. V. et al. Quantitative differences in TAD border strength underly the TAD hierarchy in Drosophila chromosomes. J. Cell. Biochem. 120, 4494–4503 (2019).
Ulianov, S. V. et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26, 70–84 (2016).
Rowley, M. J. et al. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852 (2017).
Hug, C. B., Grimaldi, A. G., Kruse, K. & Vaquerizas, J. M. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169, 216–228 (2017).
Okhovat, M. et al. TAD evolutionary and functional characterization reveals diversity in mammalian TAD boundary properties and function. Nat. Commun. 14, 8111 (2023).
Kaushal, A. et al. CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nat. Commun. 12, 1011 (2021).
Nora, E. P. et al. Molecular basis of CTCF binding polarity in genome folding. Nat. Commun. 11, 5612 (2020).
Kamalyan, S. et al. The N-terminal dimerization domains of human and Drosophila CTCF have similar functionality. Epigenetics Chromatin 17, 9 (2024).
Mohana, G. et al. Chromosome-level organization of the regulatory genome in the Drosophila nervous system. Cell 186, 3826–3844 (2023).
Zhang, H. et al. CTCF and R-loops are boundaries of cohesin-mediated DNA looping. Mol. Cell 83, 2856–2871 (2023).
Haarhuis, J. H. I. et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707 (2017).
Eagen, K. P., Aiden, E. L. & Kornberg, R. D. Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc. Natl Acad. Sci. USA 114, 8764–8769 (2017).
Li, X. et al. GAGA-associated factor fosters loop formation in the Drosophila genome. Mol. Cell 83, 1519–1526 (2023).
Batut, P. J. et al. Genome organization controls transcriptional dynamics during development. Science 375, 566–570 (2022).
Denaud, S. et al. A PRE loop at the dac locus acts as a topological chromatin structure that restricts and specifies enhancer–promoter communication. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-024-01375-7 (2024).
Ghavi-Helm, Y. et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014).
Espinola, S. M. et al. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 53, 477–486 (2021).
Pollex, T. et al. Enhancer–promoter interactions become more instructive in the transition from cell-fate specification to tissue differentiation. Nat. Genet. 56, 686–696 (2024).
modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Bonev, B. et al. Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572 (2017).
Franke, M. et al. CTCF knockout in zebrafish induces alterations in regulatory landscapes and developmental gene expression. Nat. Commun. 12, 5415 (2021).
Niu, L. et al. Three-dimensional folding dynamics of the Xenopus tropicalis genome. Nat. Genet. 53, 1075–1087 (2021).
Pérez-Rico, Y. A., Barillot, E. & Shkumatava, A. Demarcation of topologically associating domains is uncoupled from enriched CTCF binding in developing zebrafish. iScience 23, 101046 (2020).
Ortabozkoyun, H. et al. Members of an array of zinc-finger proteins specify distinct Hox chromatin boundaries. Mol. Cell 84, 3406–3422.e6 (2024).
Symmons, O. et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 24, 390–400 (2014).
Rodriguez-Carballo, E. et al. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31, 2264–2281 (2017).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene–enhancer interactions. Cell 161, 1012–1025 (2015).
Williamson, I. et al. Developmentally regulated Shh expression is robust to TAD perturbations. Development 146, dev179523 (2019).
Ghavi-Helm, Y. et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51, 1272–1282 (2019).
Despang, A. et al. Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51, 1263–1271 (2019).
Galupa, R. et al. Inversion of a topological domain leads to restricted changes in its gene expression and affects interdomain communication. Development 149, dev200568 (2022).
Galupa, R. et al. A conserved noncoding locus regulates random monoallelic Xist expression across a topological boundary. Mol. Cell 77, 352–367 (2020).
Chakraborty, S. et al. Enhancer–promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness. Nat. Genet. 55, 280–290 (2023).
Hsieh, T. S. et al. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539–553 (2020).
Hsieh, T.-H. S. et al. Enhancer–promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat. Genet. 54, 1919–1932 (2022).
Krietenstein, N. et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell 78, 554–565 (2020).
Goel, V. Y., Huseyin, M. K. & Hansen, A. S. Region capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments. Nat. Genet. 55, 1048–1056 (2023).
Roayaei Ardakany, A., Gezer, H. T., Lonardi, S. & Ay, F. Mustache: multi-scale detection of chromatin loops from Hi-C and Micro-C maps using scale-space representation. Genome Biol. 21, 256 (2020).
Benabdallah, N. S. et al. Decreased enhancer–promoter proximity accompanying enhancer activation. Mol. Cell 76, 473–484 (2019).
Zuin, J. et al. Nonlinear control of transcription through enhancer–promoter interactions. Nature 604, 571–577 (2022).
Xiao, J. Y., Hafner, A. & Boettiger, A. N. How subtle changes in 3D structure can create large changes in transcription. eLife 10, e64320 (2021).
Friman, E. T., Flyamer, I. M., Marenduzzo, D., Boyle, S. & Bickmore, W. A. Ultra-long-range interactions between active regulatory elements. Genome Res. 33, 1269–1283 (2023).
Beagrie, R. A. et al. Multiplex-GAM: genome-wide identification of chromatin contacts yields insights overlooked by Hi-C. Nat. Methods 20, 1037–1047 (2023).
Schloissnig, S. et al. The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proc. Natl Acad. Sci. USA 118, e2017176118 (2021).
Nand, A. et al. Genetic and spatial organization of the unusual chromosomes of the dinoflagellate Symbiodinium microadriaticum. Nat. Genet. 53, 618–629 (2021).
Marinov, G. K. et al. Transcription-dependent domain-scale three-dimensional genome organization in the dinoflagellate Breviolum minutum. Nat. Genet. 53, 613–617 (2021).
Janouškovec, J. et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl Acad. Sci. USA 114, E171–E180 (2017).
Kapusta, A., Suh, A. & Feschotte, C. Dynamics of genome size evolution in birds and mammals. Proc. Natl Acad. Sci. USA 114, E1460–E1469 (2017).
Lu, Z. et al. The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 5, 1250–1259 (2019).
Wu, J. et al. Systematic analysis of intron size and abundance parameters in diverse lineages. Sci. China Life Sci. 56, 968–974 (2013).
Wendel, J. F. et al. Intron size and genome size in plants. Mol. Biol. Evol. 19, 2346–2352 (2002).
Noack, F. et al. Multimodal profiling of the transcriptional regulatory landscape of the developing mouse cortex identifies Neurog2 as a key epigenome remodeler. Nat. Neurosci. 25, 154–167 (2022).
Madani Tonekaboni, S. A., Haibe-Kains, B. & Lupien, M. Large organized chromatin lysine domains help distinguish primitive from differentiated cell populations. Nat. Commun. 12, 499 (2021).
Zhang, M., Huang, H., Li, J. & Wu, Q. ZNF143 deletion alters enhancer/promoter looping and CTCF/cohesin geometry. Cell Rep. 43, 113663 (2024).
Özdemir, I. & Gambetta, M. C. The role of insulation in patterning gene expression. Genes 10, 767 (2019).
Matthews, N. E. & White, R. Chromatin architecture in the fly: living without CTCF/cohesin loop extrusion?: Alternating chromatin states provide a basis for domain architecture in Drosophila. Bioessays 41, e1900048 (2019).
Hirano, T. Condensins: organizing and segregating the genome. Curr. Biol. 15, R265–R275 (2005).
Haering, C. H., Löwe, J., Hochwagen, A. & Nasmyth, K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 (2002).
Yatskevich, S., Rhodes, J. & Nasmyth, K. Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019).
Ganji, M. et al. Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018).
Kim, E., Barth, R. & Dekker, C. Looping the genome with SMC complexes. Annu. Rev. Biochem. 92, 15–41 (2023).
Barutcu, A. R., Blencowe, B. J. & Rinn, J. L. Differential contribution of steady‐state RNA and active transcription in chromatin organization. EMBO Rep. 20, e48068 (2019).
Ing-Simmons, E. et al. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat. Genet. 53, 487–499 (2021).
Dequeker, B. J. H. et al. MCM complexes are barriers that restrict cohesin-mediated loop extrusion. Nature 606, 197–203 (2022).
Saldaña-Meyer, R. et al. RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell 76, 412–422 (2019).
Li, Y. et al. The structural basis for cohesin–CTCF-anchored loops. Nature 578, 472–476 (2020).
de Wit, E. et al. CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015).
Huang, H. et al. CTCF mediates dosage- and sequence-context-dependent transcriptional insulation by forming local chromatin domains. Nat. Genet. 53, 1064–1074 (2021).
Zhang, D. et al. Alteration of genome folding via contact domain boundary insertion. Nat. Genet. 52, 1076–1087 (2020).
Gómez-Díaz, E. & Corces, V. G. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 24, 703–711 (2014).
Kraft, K. et al. Polycomb-mediated genome architecture enables long-range spreading of H3K27 methylation. Proc. Natl Acad. Sci. USA 119, e2201883119 (2022).
Acknowledgements
D.M.I. was supported by funding from the DFG SPP 22.02 ‘3D Genome Architecture in Development and Disease’ (IB139/1-1 and IB139/6-1). Research in the Ibrahim laboratory is supported by ERC starting grant 101076709 ‘SYNREG’. M.-F.S., B.M. and G.C. were supported through the Marie Skłodowska-Curie Innovative Training Network (813327 ‘ChromDesign’ and 813282 ‘PEP-NET’) under the European Union’s Horizon 2020 research and innovation program. I.J. was supported by EMBO long-term fellowship ATLF 559-2018. G.C. was additionally supported by grants from the European Research Council (Advanced Grant 3DEpi, under grant agreement number 788972), the Fondation pour la Recherche Médicale (EQU202303016280), the MSDAVENIR foundation (project EpiMuM-3D), the Centre National pour la Recherche Scientifique, the Agence Nationale de la Recherche (‘PLASMADIFF3D’ project, ANR-18-CE15-0010) and by the French National Cancer Institute (INCa PLBIO18-362).
Author information
Authors and Affiliations
Contributions
All authors contributed substantially to discussion of the content and reviewed the literature. M.-F.S., B.M. and D.M.I. wrote an initial draft. I.J., D.M.I. and G.C. reviewed, revised and/or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Maria Cristina Gambetta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Eytan Zlotorynski and Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Szalay, MF., Majchrzycka, B., Jerković, I. et al. Evolution and function of chromatin domains across the tree of life. Nat Struct Mol Biol 31, 1824–1837 (2024). https://doi.org/10.1038/s41594-024-01427-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-024-01427-y
This article is cited by
-
The application of irreversible genomic states to define and trace ancient cell type homologies
EvoDevo (2025)
-
Three-dimensional genome architecture connects chromatin structure and function in a major wheat pathogen
BMC Biology (2025)
-
CiFi: accurate long-read chromosome conformation capture with low-input requirements
Nature Communications (2025)
-
The evolutionary foundations of transcriptional regulation in animals
Nature Reviews Genetics (2025)