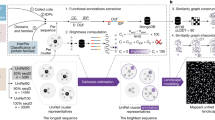
Abstract
Accurate prediction of protein structures is essential for understanding biological functions and guiding biomedical research. However, maintaining synchronization between structure models and rapidly expanding, continuously evolving protein sequence databases remains a major challenge. Here, we present AlphaSync (alphasync.stjude.org), a comprehensive resource that complements the AlphaFold Protein Structure Database. AlphaSync currently provides 2.6 million UniProt-synchronized structural models, including predictions for 40,016 updated proteins and isoforms from 925 species. AlphaSync achieves complete, up-to-date proteome coverage for 42 species, including humans, key pathogens and model organisms. It also provides residue-level annotations such as solvent accessibility, dihedral angles, intrinsic disorder status and over 4.7 billion atom-level noncovalent contacts. Its up-to-date structural models and detailed annotations will facilitate the study of protein structure–function relationships, assessment of sequence variants and machine learning tasks including protein design. With an intuitive web interface and application programming interface, AlphaSync enables protein research at scale and in detail.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Newly predicted updated structures for the latest UniProt release can be obtained online (https://alphasync.stjude.org/download under a CC BY 4.0 license). Also available for download are PAE scores and the exact AlphaFold parameters used to predict structures. AlphaSync’s residue-level data can be programmatically retrieved through the AlphaSync API (https://alphasync.stjude.org/api).
Code availability
AlphaSync’s source code is available from GitHub (https://github.com/langbnj/alphasync) under a BSD-3-Clause license. The website code is available upon request.
References
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2024).
UniProt, C. UniProt: the Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 53, D609–D617 (2025).
Bowler-Barnett, E. H. et al. UniProt and mass spectrometry-based proteomics—a 2-way working relationship. Mol. Cell Proteom. 22, 100591 (2023).
Landrum, M. J. et al. ClinVar: updates to support classifications of both germline and somatic variants. Nucleic Acids Res. 53, D1313–D1321 (2025).
Sondka, Z. et al. COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 52, D1210–D1217 (2024).
Woolard, J. et al. VEGF165b, an inhibitory vascular endothelial growth factor splice variant: mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res. 64, 7822–7835 (2004).
Tunyasuvunakool, K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021).
Lipman, D. J. & Pearson, W. R. Rapid and sensitive protein similarity searches. Science 227, 1435–1441 (1985).
Buel, G. R. & Walters, K. J. Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 29, 1–2 (2022).
Akdel, M. et al. A structural biology community assessment of AlphaFold2 applications. Nat. Struct. Mol. Biol. 29, 1056–1067 (2022).
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013).
Kayikci, M. et al. Visualization and analysis of non-covalent contacts using the Protein Contacts Atlas. Nat. Struct. Mol. Biol. 25, 185–194 (2018).
Sehnal, D. et al. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 49, W431–W437 (2021).
Harrison, P. W. et al. Ensembl 2024. Nucleic Acids Res. 52, D891–D899 (2024).
Sommer, M. J. et al. Structure-guided isoform identification for the human transcriptome. eLife 11, e82556 (2022).
Yang, Y. et al. ASpdb: an integrative knowledgebase of human protein isoforms from experimental and AI-predicted structures. Nucleic Acids Res. 53, D331–D339 (2025).
Bryant, P. & Noe, F. Structure prediction of alternative protein conformations. Nat. Commun. 15, 7328 (2024).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Kim, W. et al. Rapid and sensitive protein complex alignment with Foldseek-Multimer. Nat. Methods 22, 469–472 (2025).
Steinegger, M. et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20, 473 (2019).
Chen, X., Zaro, J. L. & Shen, W. C. Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev. 65, 1357–1369 (2013).
Joosten, R. P. et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 39, D411–D419 (2011).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Tien, M. Z., Meyer, A. G., Sydykova, D. K., Spielman, S. J. & Wilke, C. O. Maximum allowed solvent accessibilities of residues in proteins. PLoS ONE 8, e80635 (2013).
Levy, E. D. A simple definition of structural regions in proteins and its use in analyzing interface evolution. J. Mol. Biol. 403, 660–670 (2010).
Hamelryck, T. & Manderick, B. PDB file parser and structure class implemented in Python. Bioinformatics 19, 2308–2310 (2003).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Acknowledgements
We thank I. Chen, M. Phelps and D. Malinverni for their helpful input, C. Burdyshaw for help deploying AlphaFold and other members of the M.M.B. group for their helpful input and discussions. We acknowledge and thank the American Lebanese Syrian Associated Charities for financial support. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
B.L. conceptualized, built and implemented the project and wrote the manuscript. B.M. created the algorithm to predict disordered residues using AlphaFold-derived calculations and provided unpublished code and implementation examples for the disorder predictor feature. B.I.S. created the Lahuta Python package for the analysis of noncovalent interactions and protein contacts, provided access to the unpublished software for the intraprotein contact calculation feature and is helping to maintain the database. J.P. helped implement the Mol* protein structure viewer and PAE visualization. M.M.B. oversaw the project and provided guidance on the writing and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Sara Osman and Melina Casadio, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Table 1
Species included in AlphaSync and their degree of completion. This table shows the 925 species with structures available in AlphaSync, as well as the number of structures available and those still missing from its complete reviewed and canonical reference proteome. It also clearly marks the 48 model organisms and global health proteomes selected in the AFDB that we prioritized, as well as the 42 fully completed species (including isoforms) in AlphaSync.
Supplementary Table 2
AlphaFold 2 parameters used for structure prediction. This table shows an overview of the parameters, software and database versions used for AlphaFold 2 structure prediction in AlphaSync.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lang, B., Mészáros, B., Sejdiu, B.I. et al. AlphaSync is an enhanced AlphaFold structure database synchronized with UniProt. Nat Struct Mol Biol 32, 2628–2632 (2025). https://doi.org/10.1038/s41594-025-01719-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01719-x
This article is cited by
-
The aging of the AlphaFold database
Nature Structural & Molecular Biology (2025)