Abstract
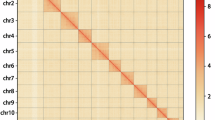
Phytophthora capsici is a globally widespread oomycete pathogen that causes severe damage to diverse host plants, posing major agricultural challenges. Here, we present a high-quality, chromosome-level genome assembly of P. capsici isolate BYA5 using PacBio HiFi, ultralong Oxford Nanopore, and Hi-C sequencing data. The assembled genome spans 83.96 Mb and comprises 18 chromosomes, capturing all centromeres and most telomeres (32/36). The assembled genome exhibits a contiguous N50 of 3.84 Mb, representing the first chromosome-level genome sequence of a pathogenic P. capsici. A total of 15,688 protein-coding genes were annotated, including 337 RxLRs and 115 Crinkler (CRN) effectors. This high-quality reference genome provides a valuable resource for advancing our understanding of oomycete pathobiology and evolution, facilitating further research on virulence mechanisms and host-pathogen interactions.
Similar content being viewed by others
Data availability
The authors confirm that the data supporting the findings of this study are available in the National Genomics Data Center (NGDC), and National Center for Biotechnology Information (NCBI), and figshare, can be accessed via. https://ngdc.cncb.ac.cn/gwh/Assembly/92621/show (assembled genome in NGDC). https://ngdc.cncb.ac.cn/gsa/browse/CRA025046 (raw data in NGDC). https://identifiers.org/ncbi/insdc.gca:GCA_053541045.1 (assembled genome in NCBI). https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1248058 (raw data in NCBI). https://doi.org/10.6084/m9.figshare.28856261 (genome annotations archive in figshare).
Code availability
All commands and pipelines used in data processing were executed according to the manual and protocols of the corresponding bioinformatic software and described in the Methods section, along with the versions. No custom code was generated for these analyses.
References
Lamour, K. H., Stam, R., Jupe, J. & Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Molecular Plant Pathology 13, 329–337, https://doi.org/10.1111/j.1364-3703.2011.00754.x (2012).
Kamoun, S. et al. The Top 10 oomycete pathogens in molecular plant pathology. Molecular Plant Pathology 16, 413–434, https://doi.org/10.1111/mpp.12190 (2015).
Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review Of Phytopathology 44, 41–60, https://doi.org/10.1146/annurev.phyto.44.070505.143436 (2006).
Białas, A. et al. Lessons in Effector and NLR Biology of Plant-Microbe Systems. Molecular Plant-Microbe Interactions: MPMI 31, 34–45, https://doi.org/10.1094/MPMI-08-17-0196-FI (2018).
Anderson, R. G., Deb, D., Fedkenheuer, K. & McDowell, J. M. Recent Progress in RXLR Effector Research. Molecular Plant-Microbe Interactions: MPMI 28, 1063–1072, https://doi.org/10.1094/MPMI-01-15-0022-CR (2015).
Birch, P. R. J., Rehmany, A. P., Pritchard, L., Kamoun, S. & Beynon, J. L. Trafficking arms: oomycete effectors enter host plant cells. Trends in Microbiology 14, 8–11, https://doi.org/10.1016/j.tim.2005.11.007 (2006).
Zhang, Z. et al. Complete telomere-to-telomere genomes uncover virulence evolution conferred by chromosome fusion in oomycete plant pathogens. Nature Communications 15, 4624, https://doi.org/10.1038/s41467-024-49061-y (2024).
Tsykun, T. et al. High-Quality Genome of the Tree Pathogen Phytophthora plurivora—A Novel Resource for Epidemiological Research. PhytoFrontiersTM 3, 888–892, https://doi.org/10.1094/PHYTOFR-05-23-0065-A (2023).
Matson, M. E. H., Liang, Q., Lonardi, S. & Judelson, H. S. Karyotype variation, spontaneous genome rearrangements affecting chemical insensitivity, and expression level polymorphisms in the plant pathogen Phytophthora infestans revealed using its first chromosome-scale assembly. PLOS Pathogens 18, e1010869, https://doi.org/10.1371/journal.ppat.1010869 (2022).
Cox, M. P. et al. Chromosome-level assembly of the Phytophthora agathidicida genome reveals adaptation in effector gene families. Frontiers in Microbiology 13, 1038444, https://doi.org/10.3389/fmicb.2022.1038444 (2022).
Lamour, K. H. et al. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Molecular Plant-Microbe Interactions: MPMI 25, 1350–1360, https://doi.org/10.1094/MPMI-02-12-0028-R (2012).
Reyes-Tena, A. et al. Genome Sequence Data of Six Isolates of Phytophthora capsici from Mexico. Molecular Plant-Microbe Interactions: MPMI 32, 1267–1269, https://doi.org/10.1094/MPMI-01-19-0014-A (2019).
Lee, J. H., Siddique, M. I., Kwon, J. K. & Kang, B.-C. Comparative Genomic Analysis Reveals Genetic Variation and Adaptive Evolution in the Pathogenicity-Related Genes of Phytophthora capsici. Frontiers in Microbiology 12, 69413, https://doi.org/10.3389/fmicb.2021.694136 (2021).
Villanueva, O., Nguyen, H. D. T. & Ellouze, W. Comparative Genomic and Secretome Analysis of Phytophthora capsici Strains: Exploring Pathogenicity and Evolutionary Dynamics. Agronomy 14, 2623, https://doi.org/10.3390/agronomy14112623 (2024).
Szadkowski, E. et al. Phytophthora capsici genome assembly for two isolates using long-read Oxford Nanopore Technology sequencing. Microbiology Resource Announcements 12, e0019623, https://doi.org/10.1128/MRA.00196-23 (2023).
Cui, C., Herlihy, J. H., Bombarely, A., McDowell, J. M. & Haak, D. C. Draft Assembly of Phytophthora capsici from Long-Read Sequencing Uncovers Complexity. Molecular Plant-Microbe Interactions: MPMI 32, 1559–1563, https://doi.org/10.1094/MPMI-04-19-0103-TA (2019).
Shi, J. et al. Improved Whole-Genome Sequence of Phytophthora capsici Generated by Long-Read Sequencing. Molecular Plant-Microbe Interactions: MPMI 34, 866–869, https://doi.org/10.1094/MPMI-12-20-0356-A (2021).
A, J. et al. Pathogenomics Insights into Phytophthora capsici and Phytophthora tropicalis-Sibling Species Causing Black Pepper Foot Rot: Genomic Architecture. Metabolic Pathways, and Effector Diversity. Gene 947, 149328, https://doi.org/10.1016/j.gene.2025.149328 (2025).
Wang, W. et al. Functional Analysis of the C-5 Sterol Desaturase PcErg3 in the Sterol Auxotrophic Oomycete Pathogen Phytophthora capsici. Frontiers in Microbiology 10, 13, https://doi.org/10.3389/fmicb.2022.811132 (2022).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods 18, 170–175, https://doi.org/10.1038/s41592-020-01056-5 (2021).
Winkworth, R. et al. Comparative Analyses of Complete Peronosporaceae (Oomycota) Mitogenome Sequences-Insights into Structural Evolution and Phylogeny. Genome Biology and Evolution 14, evac049, https://doi.org/10.1093/gbe/evac049 (2022).
Zhang, X., Zhang, S., Zhao, Q., Ming, R. & Tang, H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nature Plants 5, 833–845, https://doi.org/10.1038/s41477-019-0487-8 (2019).
Durand, N. C. et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Systems 3, 99–101, https://doi.org/10.1016/j.cels.2015.07.012 (2016).
Jain, M., Olsen, H. E., Paten, B. & Akeson, M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biology 17, 239, https://doi.org/10.1186/s13059-016-1103-0 (2016).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biology 21, 245, https://doi.org/10.1186/s13059-020-02134-9 (2020).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Research 34, W435–439, https://doi.org/10.1093/nar/gkl200 (2006).
Wang, Y., Pruitt, R. N., Nürnberger, T. & Wang, Y. Evasion of plant immunity by microbial pathogens. Nature Reviews Microbiology 20, 449–464, https://doi.org/10.1038/s41579-022-00710-3 (2022).
Wilson, R. A. & McDowell, J. M. Recent advances in understanding of fungal and oomycete effectors. Current Opinion In Plant Biology 68, 102228, https://doi.org/10.1016/j.pbi.2022.102228 (2022).
Fernandez, J. The Phantom Menace: latest findings on effector biology in the rice blast fungus. aBIOTECH 4, 140–154, https://doi.org/10.1007/s42994-023-00099-4 (2023).
Zheng, Z. et al. Atypical RXLR effectors are involved in Phytophthora cactorum pathogenesis. aBIOTECH 6, 50–62, https://doi.org/10.1007/s42994-025-00198-4 (2025).
Miao, J., Chi, Y., Lin, D., Tyler, B. M. & Liu, X. Mutations in ORP1 Conferring Oxathiapiprolin Resistance Confirmed by Genome Editing using CRISPR/Cas9 in Phytophthora capsici and P. sojae. Phytopathology 108, 1412–1419, https://doi.org/10.1094/PHYTO-01-18-0010-R (2018).
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S. & Thompson, W. F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature Protocols 1, 2320–2325, https://doi.org/10.1038/nprot.2006.384 (2006).
Belton, J. M. et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods San Diego Calif 58, 268–276, https://doi.org/10.1016/j.ymeth.2012.05.001 (2012).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770, https://doi.org/10.1093/bioinformatics/btr011 (2011).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890, https://doi.org/10.1093/bioinformatics/bty560 (2018).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 37, 907–915, https://doi.org/10.1038/s41587-019-0201-4 (2019).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. PNAS 117, 9451–9457, https://doi.org/10.1073/pnas.1921046117 (2020).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Research 14, 988–995, https://doi.org/10.1101/gr.1865504 (2004).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature Protocols 11, 1650–1667, https://doi.org/10.1038/nprot.2016.095 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29, 644–652, https://doi.org/10.1038/nbt.1883 (2011).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biology 9, R7, https://doi.org/10.1186/gb-2008-9-1-r7 (2008).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Research 33, D121–124, https://doi.org/10.1093/nar/gki081 (2005).
Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Research 49, 9077–9096, https://doi.org/10.1093/nar/gkab688 (2021).
Sperschneider, J. & Dodds, P. N. EffectorP 3.0: Prediction of Apoplastic and Cytoplasmic Effectors in Fungi and Oomycetes. Molecular Plant-Microbe Interactions. MPMI 35, 146–156, https://doi.org/10.1094/mpmi-08-21-0201-r (2022).
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340, 783–795, https://doi.org/10.1016/j.jmb.2004.05.028 (2004).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580, https://doi.org/10.1006/jmbi.2000.4315 (2001).
Wan, Y. This study aimed to obtain high quality genomic sequence of Phytophthora capsica isolate BYA5. BioProject https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1248058 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR33296427 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR33296426 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR33296420 (2025).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_053541045.1 (2025).
NGDC Genome Sequence Archive https://ngdc.cncb.ac.cn/gsa/browse/CRA025046 (2025).
NGDC Genome Sequence Archive https://ngdc.cncb.ac.cn/gwh/Assembly/92621/show (2025).
Wan, Y. The annotated file for Phytophthora capsici isolate BYA5. Figshare https://doi.org/10.6084/m9.figshare.28856261 (2025).
Lin, Y. et al. quarTeT: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Horticulture Research 10, uhad127, https://doi.org/10.1093/hr/uhad127 (2023).
Brown, M. R., Manuel Gonzalez de La Rosa, P. & Blaxter, M. tidk: a toolkit to rapidly identify telomeric repeats from genomic datasets. Bioinformatics 41, btaf049, https://doi.org/10.1093/bioinformatics/btaf049 (2025).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, https://doi.org/10.1093/bioinformatics/btp324 (2009).
Acknowledgements
This work was supported by Taishan Scholar Foundation of Shandong Province (tsqn202103162, tsqn202211093), Key R&D Program of Shandong Province, China (2024CXPT03), Yunnan Department of Science and Technology-Development and adoption of innovative ‘green’ management tools against insect pest and disease on tomato and pepper in Yunnan (No. 202502AQ370001), the Natural Science Foundation of Shandong Province (SYS202206), National Natural Science Foundation of China (32172387). We thank Dr. Xili Liu of Northwest Agriculture and Forestry University for providing P. capsici isolate BYA5. We thank Dr. Daolong Dou of Nanjing Agricultural University for providing the genomic data and GFF annotation files of P. capsici isolate LT263. This work was supported by the bioinformatics services from Novogene Bioinformatics Technology Co., Ltd. (Tianjin, China), particularly in data processing and genomic annotation.
Author information
Authors and Affiliations
Contributions
Y.C. and Q.W. conceived and supervised the project. Y.W., F.Z., C.X. and X.L. collected the samples and strain preservation; M.Z. and Y.W. performed the genomic data analysis; S.G. submitted the genome assembly data to the NCBI database. Y.W. and F.Z. drafted the initial version of the manuscript. Q.W. and Y.C. finalized the manuscript. All authors read and prove the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wan, Y., Zhu, F., Zhang, M. et al. Chromosome-scale assembly and annotation of Phytophthora capsici isolate BYA5. Sci Data (2026). https://doi.org/10.1038/s41597-025-06501-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-06501-8