Abstract
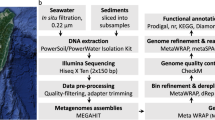
Actively venting high temperature deep-sea hydrothermal vent deposits along tectonic spreading centers and in backarc basins harbor a rich diversity of thermophilic Bacteria and Archaea, many of which have no representatives in cultivation nor any genomic representation in databases. Here, in order to produce a global-scale time series metagenomic resource for studying the microbial functional and genomic diversity in these high temperature ecosystems, we obtained 70 metagenomes from collections across spatial and temporal gradients from 21 different vent fields spanning 16 years (1993–2009). The dataset (Deep-Sea Hydrothermal Vent dataset (DSV70)) includes 3.56 Tbp of raw DNA sequence reads, that have been assembled to produce 7,422 medium- to high-quality (based on CheckM2) metagenome-assembled genomes (MAGs) of Bacteria (6,063 MAGs) and Archaea (1,359 MAGs). Collectively, this DSV70 dataset and the published 40 metagenomes from more recent deep-sea collections (2004 to 2018), represent a valuable resource for exploring the functional and phylogenomic diversity of the deep-sea hydrothermal microbiomes, and provide many reference genomes for studies in the taxonomy and systematics of poorly studied microbial lineages. Further, with the interest in mining the mineral resources at deep-sea vents, the DSV70 provides a genomic legacy for monitoring impacts on the microbial communities in these systems.
Similar content being viewed by others
Data availability
The primary metagenome sequences and the MAGs of ≥90% completion have been deposited in GenBank and the NCBI Sequence Read Archive (SRA), respectively, under PRJNA1244896 and SRP575724. Primary metagenome sequences, metagenome assemblies and all medium- to high-quality MAGs are available in the European Bioinformatics Institute BioStudies repository (accession number S-BSST2227).
Code availability
All of the bioinformatic tools presented in this analysis are publicly available, and version numbers and parameters are described in detail in the Methods. Custom scripts for calculating normalized MAG coverage are available at https://github.com/ALR-Lab/MAG-coverage.
References
Tivey, M. K. Generation of seafloor hydrothermal vent fluids and associated mineral deposits. Oceanography 20, 50–65, https://doi.org/10.5670/oceanog.2007.80 (2007).
Flores, G. E. et al. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 13, 2158–2171, https://doi.org/10.1111/j.1462-2920.2011.02463.x (2011).
Flores, G. E. et al. Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology 10, 333–346, https://doi.org/10.1111/j.1472-4669.2012.00325.x (2012).
Reysenbach, A.-L. et al. Complex subsurface hydrothermal fluid mixing at a submarine arc volcano supports distinct and highly diverse microbial communities. Proc. Natl Acad. Sci. USA 117, 32627–32638, https://doi.org/10.1073/pnas.2019021117 (2020).
Zhou, Z., St. John, E., Anantharaman, K. & Reysenbach, A.-L. Global patterns of diversity and metabolism of microbial communities in deep-sea hydrothermal vent deposits. Microbiome 10, 241, https://doi.org/10.1186/s40168-022-01424-7 (2022).
Langwig, M. V. et al. Endemism shapes viral ecology and evolution in globally distributed hydrothermal vent ecosystems. Nat. Commun. 16, 4076, https://doi.org/10.1038/s41467-025-59154-x (2025).
Gruber-Vodicka, H. R., Seah, B. K. B. & Pruesse, E. phyloFlash: rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems 5, e00920-20, https://doi.org/10.1128/msystems.00920-20 (2020).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119, https://doi.org/10.1186/1471-2105-11-119 (2010).
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34, https://doi.org/10.1093/nar/27.1.29 (1999).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731, https://doi.org/10.1016/j.jmb.2015.11.006 (2016).
Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158, https://doi.org/10.1186/s40168-018-0541-1 (2018).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927, https://doi.org/10.1093/bioinformatics/btz848 (2020).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004, https://doi.org/10.1038/nbt.4229 (2018).
Rinke, C. et al. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 6, 946–959, https://doi.org/10.1038/s41564-021-00918-8 (2021).
GenBank https://identifiers.org/ncbi/bioproject:PRJNA1244896 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP575724 (2025).
St. John, E. & Reysenbach, A.-L. Data records for “Global deep-sea hydrothermal deposit metagenomes and metagenome-assembled genomes over time and space”. BioStudies Database https://identifiers.org/biostudies:S-BSST2227 (2025).
Qi, Y.-L., Zhang, H.-T., Li, M., Li, W.-J. & Hua, Z.-S. Recovery of nearly 3,000 archaeal genomes from 152 terrestrial geothermal spring metagenomes. Sci Data 12, 151, https://doi.org/10.1038/s41597-025-04493-z (2025).
Sunagawa, S. et al. Tara Oceans: towards global ocean ecosystems biology. Nat. Rev. Microbiol. 18, 428–445, https://doi.org/10.1038/s41579-020-0364-5 (2020).
Sánchez, P. et al. Marine picoplankton metagenomes and MAGs from eleven vertical profiles obtained by the Malaspina Expedition. Sci. Data 11, 154, https://doi.org/10.1038/s41597-024-02974-1 (2024).
Clayton, S. et al. Bio-GO-SHIP: the time is right to establish global repeat sections of ocean biology. Front. Mar. Sci. 8, 767443, https://doi.org/10.3389/fmars.2021.767443 (2022).
Biller, S. J. et al. Marine microbial metagenomes sampled across space and time. Sci. Data 5, 180176, https://doi.org/10.1038/sdata.2018.176 (2018).
Nishimura, Y. & Yoshizawa, S. The OceanDNA MAG catalog contains over 50,000 prokaryotic genomes originated from various marine environments. Sci. Data 9, 305, https://doi.org/10.1038/s41597-022-01392-5 (2022).
Chen, J. et al. Global marine microbial diversity and its potential in bioprospecting. Nature 633, 371–379, https://doi.org/10.1038/s41586-024-07891-2 (2024).
Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2, e107, https://doi.org/10.1002/imt2.107 (2023).
Bushnell, B. BBMap: a fast, accurate, splice-aware aligner. In 9th Annual Genomics of Energy & Environment Meeting (Lawrence Berkeley National Laboratory, Berkeley, CA, USA, 2014).
bbmap. Masked version of hG19 by Brian Bushnell. Zenodo https://doi.org/10.5281/zenodo.1208052 (2018).
Rodriguez-R, L. M. & Konstantinidis, K. T. Nonpareil: a redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics 30, 629–635, https://doi.org/10.1093/bioinformatics/btt584 (2014).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596, https://doi.org/10.1093/nar/gks1219 (2013).
Li, D. et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11, https://doi.org/10.1016/j.ymeth.2016.02.020 (2016).
Astashyn, A. et al. Rapid and sensitive detection of genome contamination at scale with FCS-GX. Genome Biol. 25, 60, https://doi.org/10.1186/s13059-024-03198-7 (2024).
The Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 52, W83–W94, https://doi.org/10.1093/nar/gkae410 (2024).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359, https://doi.org/10.1038/nmeth.1923 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079, https://doi.org/10.1093/bioinformatics/btp352 (2009).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359, https://doi.org/10.7717/peerj.7359 (2019).
Wu, Y.-W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607, https://doi.org/10.1093/bioinformatics/btv638 (2016).
Kang, D. D., Froula, J., Egan, R. & Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165, https://doi.org/10.7717/peerj.1165 (2015).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055, https://doi.org/10.1101/gr.186072.114 (2015).
Chklovski, A., Parks, D. H., Woodcroft, B. J. & Tyson, G. W. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 20, 1203–1212, https://doi.org/10.1038/s41592-023-01940-w (2023).
Chang, T., Gavelis, G. S., Brown, J. M. & Stepanauskas, R. Genomic representativeness and chimerism in large collections of SAGs and MAGs of marine prokaryoplankton. Microbiome 12, 126, https://doi.org/10.1186/s40168-024-01848-3 (2024).
Orakov, A. et al. GUNC: detection of chimerism and contamination in prokaryotic genomes. Genome Biol. 22, 178, https://doi.org/10.1186/s13059-021-02393-0 (2021).
Parks, D. H. et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1079–1086, https://doi.org/10.1038/s41587-020-0501-8 (2020).
von Meijenfeldt, F. A. B., Arkhipova, K., Cambuy, D. D., Coutinho, F. H. & Dutilh, B. E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 20, 217, https://doi.org/10.1186/s13059-019-1817-x (2019).
Rodriguez-R, L. M. et al. An ANI gap within bacterial species that advances the definitions of intra-species units. mBio 15, e02696-23, https://doi.org/10.1128/mbio.02696-23 (2024).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868, https://doi.org/10.1038/ismej.2017.126 (2017).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490, https://doi.org/10.1371/journal.pone.0009490 (2010).
Shimodaira, H. & Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116, https://doi.org/10.1093/oxfordjournals.molbev.a026201 (1999).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82, https://doi.org/10.1093/nar/gkae268 (2024).
GEBCO Compilation Group. GEBCO 2024 Grid, https://doi.org/10.5285/1c44ce99-0a0d-5f4f-e063-7086abc0ea0f (2024).
Coffin, M. F., Gahagan, L. M. & Lawver, L. A. Present-day plate boundary digital data compilation. University of Texas Institute for Geophysics Technical Report 174 (1997).
Acknowledgements
Thank-you to the crews of the R/V Atlantis, R/V Knorr, R/V Le Nadir, R/V Melville, R/V Roger Revelle, R/V Thomas G. Thompson, R/V Western Flyer, R/V Yokosuka, DSV Alvin, DSV Nautile, DSV Shinkai 6500, ROV Jason I, Jason II and ROV Tiburon for their help with sample collection. Our thanks to Jennifer Meneghin for her assistance in developing coverage-related scripts. This work was funded by the US-National Science Foundation grant DEB-2409507 to A.-L. Reysenbach. The US-National Science Foundation grants OCE-1558795, 1235432, 0937404, 0752469, 0728391, 0242038, 0118240, 9714302 provided funding for the many collections.
Author information
Authors and Affiliations
Contributions
E.S.J. extracted and quantified DNA, conducted the metagenome trimming, quality control, assembly, read mapping and binning, phylogenomic tree construction and figure generation, collected metadata and helped write the manuscript. A.L.R. designed the study, collected the samples, helped analyze the data and wrote the manuscript. Both authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
St. John, E., Reysenbach, AL. Global deep-sea hydrothermal deposit metagenomes and metagenome-assembled genomes over time and space. Sci Data (2026). https://doi.org/10.1038/s41597-026-06612-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06612-w