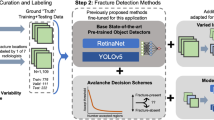
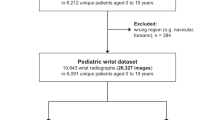
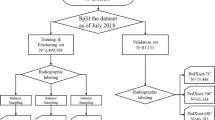
Abstract
Pediatric forearm fractures, particularly involving the ulna and radius, are among the most common childhood injuries. However, the lack of standardized and openly available datasets has limited progress in artificial intelligence research and constrained clinical validation. To address this issue, we present the Pediatric Ulna and Radius Fractures (PediURF) dataset, a first-of-its-kind, publicly available collection of over 10,000 de-identified images. Each image is carefully annotated by expert radiologists and categorized into three clinically relevant types: proximal, midshaft, and distal fractures. By releasing PediURF, we aim to provide an accessible resource for deep learning-based models development, benchmarking, and clinical training. To validate its utility, we proposed URFNet, a dual-view classification model designed to integrate anteroposterior and lateral perspectives. The proposed model achieved the best performance when compared with other classification models. Collectively, the proposed PediURF dataset provides a valuable foundation for future deep learning-based studies in pediatric fracture classification.
Similar content being viewed by others
Data availability
The proposed dataset collected in this work has been fully de-identified and released under an open-access license to support reproducibility and reuse. The dataset is available at https://doi.org/10.6084/m9.figshare.29998954.
Code availability
The source code for the proposed URFNet model, together with the complete experimental framework, is openly accessible at https://github.com/GG-Tang/URFNet.
The repository includes all scripts necessary to replicate the results of this study. The main directory provides implementations for the proposed URFNet construction (models.py), dataset handling and preprocessing (datajson.py and dataset.py), as well as training (train.py) and evaluation (test.py) procedures. In addition, a dedicated folder named Comparison models contains reference implementations of baseline models, enabling direct benchmarking of URFNet against widely used convolutional and transformer-based networks.
To facilitate reproducibility, the repository also provides a requirements.txt file that lists the required Python dependencies, ensuring consistency across different computing environments. By making this implementation freely available, we aim to ensure transparency, allow independent verification of our findings, and promote further development of deep learning methods for pediatric fracture classification.
References
Shelmerdine, S. C., White, R. D., Liu, H., Arthurs, O. J. & Sebire, N. J. Artificial intelligence for radiological paediatric fracture assessment: a systematic review. Insights into Imaging 13(1), 94, https://doi.org/10.1186/s13244-022-01234-3 (2022).
Lempesis, V. et al. Time trends in pediatric fracture incidence in Sweden during the period 1950–2006. Acta orthopaedica 88(4), 440–445, https://doi.org/10.1080/17453674.2017.1334284 (2017).
Sinikumpu, J. J., Pokka, T. & Serlo, W. The changing pattern of pediatric both-bone forearm shaft fractures among 86,000 children from 1997 to 2009. European journal of pediatric surgery 23(04), 289–296, https://doi.org/10.1055/s-0032-1333116 (2013).
Qiu, X. et al. Upper limb pediatric fractures in 22 tertiary children’s hospitals, China: a multicenter epidemiological investigation and economic factor analysis of 32,832 hospitalized children. Journal of orthopaedic surgery and research 17(1), 300, https://doi.org/10.1186/s13018-022-03159-5 (2022).
Al‐Sani, F. et al. Adverse events from emergency physician pediatric extremity radiograph interpretations: a prospective cohort study. Academic Emergency Medicine 27(2), 128–138, https://doi.org/10.1111/acem.13884 (2020).
Till, T., Tschauner, S., Singer, G., Lichtenegger, K. & Till, H. Development and optimization of AI algorithms for wrist fracture detection in children using a freely available dataset. Frontiers in Pediatrics 11, 1291804, https://doi.org/10.3389/fped.2023.1291804 (2023).
Ju, R. Y. & Cai, W. Fracture detection in pediatric wrist trauma X-ray images using YOLOv8 algorithm. Scientific Reports 13(1), 20077, https://doi.org/10.1038/s41598-023-47460-7 (2023).
Zech, J. R. et al. Detecting pediatric wrist fractures using deep-learning-based object detection. Pediatric Radiology 53(6), 1125–1134, https://doi.org/10.1007/s00247-023-05588-8 (2023).
Ferdi, A. Lightweight G-YOLOv11: Advancing Efficient Fracture Detection in Pediatric Wrist X-rays. Biomedical Signal Processing and Control 113, 108816, https://doi.org/10.1016/j.bspc.2025.108861 (2025).
Zech, J. R., Jaramillo, D., Altosaar, J., Popkin, C. A. & Wong, T. T. Artificial intelligence to identify fractures on pediatric and young adult upper extremity radiographs. Pediatric Radiology 53(12), 2386–2397, https://doi.org/10.1007/s00247-023-05754-y (2023).
Ali, S. N. E., Sherif, H. M., Hassan, S. M. & El Marakby, A. A. E. R. Long bones x-ray fracture classification using machine learning. Journal of Al-Azhar University Engineering Sector 19(72), 121–133, https://doi.org/10.21608/auej.2024.259630.1577 (2024).
Alam, A. et al. Novel transfer learning based bone fracture detection using radiographic images. BMC Medical Imaging 25(1), 5, https://doi.org/10.1186/s12880-024-01546-4 (2025).
Tang, S. et al. A Comprehensive X-ray Dataset for Pediatric Ulna and Radius Fractures Analysis. figshare https://doi.org/10.6084/m9.figshare.29998954 (2025).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Communications of the ACM 60(6), 84–90, https://doi.org/10.1145/3065386 (2017).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arxiv preprint arxiv:1409.1556, https://arxiv.org/abs/1409.1556 (2014).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770-778, https://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html (2016).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 4700-4708, https://arxiv.org/abs/1608.06993 (2017).
Howard, A. G. et al Mobilenets: Efficient convolutional neural networks for mobile vision applications. arxiv preprint arxiv:1704.04861, https://arxiv.org/abs/1704.04861 (2017).
Tan, M. & Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In International conference on machine learning 97, 6105–6114, https://proceedings.mlr.press/v97/tan19a.html?ref=ji (2019).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. arxiv preprint arxiv:2010.11929, https://arxiv.org/pdf/2010.11929/1000 (2020).
Touvron, H. et al. Training data-efficient image transformers & distillation through attention. In International conference on machine learning 139, 10347–10357, https://proceedings.mlr.press/v139/touvron21a (2021).
Li, Y. et al. Efficientformer: Vision transformers at mobilenet speed. Advances in Neural Information Processing Systems 35, 12934–12949, https://arxiv.org/abs/2206.01191 (2022).
Pham, H. H., Trung, H. N. & Nguyen, H. Q. VinDr-SpineXR: A large annotated medical image dataset for spinal lesions detection and classification from radiographs. PhysioNet, RRID-SCR_007345, https://www.physionet.org/content/vindr-spinexr/1.0.0/ (2021).
Cheng, C. T. et al. A scalable physician-level deep learning algorithm detects universal trauma on pelvic radiographs. Nature communications 12(1), 1066, https://doi.org/10.1038/s41467-021-21311-3 (2021).
Nagy, E., Janisch, M., Hržić, F., Sorantin, E. & Tschauner, S. A pediatric wrist trauma x-ray dataset (grazpedwri-dx) for machine learning. Scientific data 9(1), 222, https://doi.org/10.1038/s41597-022-01328-z (2022).
Janisch, M. et al. Pediatric radius torus fractures in x-rays—how computer vision could render lateral projections obsolete. Frontiers in Pediatrics 10, 1005099, https://doi.org/10.3389/fped.2022.1005099 (2022).
Abedeen, I. et al. Fracatlas: A dataset for fracture classification, localization and segmentation of musculoskeletal radiographs. Scientific data 10(1), 521, https://doi.org/10.1038/s41597-023-02432-4 (2023).
Pham, H. H., Nguyen, N. H., Tran, T. T., Nguyen, T. N. & Nguyen, H. Q. PediCXR: an open, large-scale chest radiograph dataset for interpretation of common thoracic diseases in children. Scientific data 10(1), https://doi.org/10.1038/s41597-023-02102-5 240 (2023).
Talha, F. F. T., Jahan, M. S. & Mojumdar, M. U. X-ray Bone Fracture Dataset: Comprehensive Imaging for Fracture Classification and Medical Research. Mendeley Data, 1, https://doi.org/10.17632/8d9kn57pdj.1 (2024).
Ran, Y. et al. A high-quality dataset featuring classified and annotated cervical spine X-ray atlas. Scientific data 11(1), 625, https://doi.org/10.1038/s41597-024-03383-0 (2024).
Yari, A., Fasih, P., Hosseini Hooshiar, M., Goodarzi, A. & Fattahi, S. F. Detection and classification of mandibular fractures in panoramic radiography using artificial intelligence. Dentomaxillofacial Radiology 53(6), 363–371, https://doi.org/10.1093/dmfr/twae018 (2024).
Li, Y. et al. Proximal femur fracture detection on plain radiography via feature pyramid networks. Scientific reports 14(1), 12046, https://doi.org/10.1038/s41598-024-63001-2 (2024).
Acknowledgements
This work was supported by the GuangDong Basic and Applied Basic Research Foundation (2024A1515110113), the Dongguan University of Technology (221110211), and the Science and Technology Development Fund, Macau SAR (0096/2023/RIA2, 0123/2022/A3). This work is also supported by the Guangdong Province Graduate Education Innovation Project (2025JGXM_149).
Author information
Authors and Affiliations
Contributions
Conceptualization: S.T.; Methodology: S.T.; Software, S.T. and L.O.; Validation: S.T.; Formal analysis: S.T.; Investigation: L.O., and W.L.; Resources: Z.X. and Z.Z.; Data curation: S.T.; Writing–original draft preparation: S.T., L.O., W.L., Z.X., N.L., H.L., and Y.L.; Writing–review and editing: ST., L.O., W.L., Z.X., N.L., H.L., Y.L, and Z.Z.; Visualization, W.L., N.L., and H.L.; Supervision: S.T., Z.X. and Z.Z.; Project administration: Z.X. and Z.Z.; Funding acquisition: S.T., Y.L. and Z.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tang, S., Ou, L., Li, W. et al. A Comprehensive X-ray Dataset for Pediatric Ulna and Radius Fractures Analysis. Sci Data (2026). https://doi.org/10.1038/s41597-026-06666-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06666-w