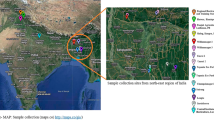
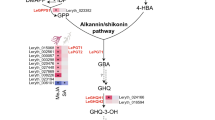
Abstract
The genus Oldenlandia encompasses numerous medicinal plants with significant pharmacological potential, yet their tissue-specific metabolic pathways and transcriptional profiles remain understudied. This study combined transcriptomic and metabolomic analyses to compare the differences in five tissues (roots, stems, leaves, flowers, and fruits) between two closely related Oldenlandia species, Oldenlandia diffusa and Oldenlandia corymbosa. Using a widely targeted metabolomics approach based on UPLC-MS/MS, we monitored 1,343 metabolites across 60 samples from the two species. Transcriptome sequencing generated 272.12 Gb and 257.03 Gb of clean data for Oldenlandia diffusa and Oldenlandia corymbosa, respectively, along with 37,644 and 20,825 annotated genes. Comparative analysis revealed numerous differentially expressed genes, whose diverse functions were elucidated through GO enrichment. This comprehensive dataset provides a valuable resource for further investigating the phenotypic traits and metabolic mechanisms underlying the pharmacological activities of these medicinally important Oldenlandia species.
Similar content being viewed by others
Data availability
The original data of the present study cohort were deposited in the NCBI public repository (https://identifiers.org/ncbi/insdc.sra:SRP665145). Metabolomic raw data have been archived in MetaboLights (MTBLS13314, https://www.ebi.ac.uk/metabolights/MTBLS13314). Additionally, expression and quality control data for metabolome and transcriptome analyses have been uploaded to the figshare database for accessibility.
Code availability
The methods section details the software and R packages employed for metabolomic and transcriptomic analyses. The R scripts for the comparative transcriptome analysis of the two species have been stored in figshare.
References
Neupane, S. et al. The Hedyotis-Oldenlandia complex (Rubiaceae: Spermacoceae) in Asia and the Pacific: Phylogeny revisited with new generic delimitations. Taxon 64, 299–322 (2015).
Al-Shuhaib, M. B. S. & Al-Shuhaib, J. M. B. Phytochemistry, pharmacology, and medical uses of Oldenlandia (family Rubaceae): a review. Naunyn Schmiedebergs Arch Pharmacol 397, 2021–2053, https://doi.org/10.1007/s00210-023-02756-3 (2024).
Lv, Y. & Wang, Y. Chemical constituents from Oldenlandia diffusa and their cytotoxic effects on human cancer cell lines. Nat Prod Res 37, 397–403, https://doi.org/10.1080/14786419.2021.1974434 (2023).
Lin, C. C. et al. Anti-inflammatory and hepatoprotective activity of peh-hue-juwa-chi-cao in male rats. Am J Chin Med 30, 225–234, https://doi.org/10.1142/s0192415x02000405 (2002).
Liuying, D. & Qiyang, J. Investigation and Identification of Varieties of Hedyotis diffusa in Fujian. Fujian Journal of Chinese Medicine, 56–58 (1982)
Qiqi, C. et al. Identification and Comparative Medicinal Progress of Hedyotis diffusa and Hedyotis corymbosa. Chinese Herbal Medicine 48, 4328–4338 (2017).
Zheng, C. C. Research on the identification of Hedyotis diffusa and its confusable species in our province. Hainan Medicine, 57–58 (1990).
Yuan, P. et al. Transcriptome regulation of carotenoids in five flesh-colored watermelons (Citrullus lanatus). BMC Plant Biol 21, 203, https://doi.org/10.1186/s12870-021-02965-z (2021).
Zhou, Y. et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat Plants 2, 16183, https://doi.org/10.1038/nplants.2016.183 (2016).
Li, R. et al. Capsaicin Attenuates Oleic Acid-Induced Lipid Accumulation via the Regulation of Circadian Clock Genes in HepG2 Cells. J Agric Food Chem 70, 794–803, https://doi.org/10.1021/acs.jafc.1c06437 (2022).
Chen, F. et al. Different accumulation profiles of multiple components between pericarp and seed of Alpinia oxyphylla capsular fruit as determined by UFLC-MS/MS. Molecules 19, 4510–4523, https://doi.org/10.3390/molecules19044510 (2014).
Gupta, S. et al. Anticancer activities of Oldenlandia diffusa. J Herb Pharmacother 4, 21–33 (2004).
Sunwoo, Y. Y. et al. Oldenlandia diffusa Promotes Antiproliferative and Apoptotic Effects in a Rat Hepatocellular Carcinoma with Liver Cirrhosis. Evid Based Complement Alternat Med 2015, 501508, https://doi.org/10.1155/2015/501508 (2015).
Wu, M., Northen, T. R. & Ding, Y. Stressing the importance of plant specialized metabolites: omics-based approaches for discovering specialized metabolism in plant stress responses. Front Plant Sci 14, 1272363, https://doi.org/10.3389/fpls.2023.1272363 (2023).
Itkin, M. et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proceedings of the National Academy of Sciences 113, E7619–E7628, https://doi.org/10.1073/pnas.1604828113 (2016).
Xiong, X. et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nat Plants 7, 1026–1036, https://doi.org/10.1038/s41477-021-00963-5 (2021).
Gao, Y, Xu, D & Hu, Z.Telomere-to-telomere genome assembly of Oldenlandia diffusa. DNA Res 31, https://doi.org/10.1093/dnares/dsae012 (2024).
Julca, I. et al. Genomic, transcriptomic, and metabolomic analysis of Oldenlandia corymbosa reveals the biosynthesis and mode of action of anti-cancer metabolites. J Integr Plant Biol 65, 1442–1466, https://doi.org/10.1111/jipb.13469 (2023).
Chen, W. et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant 6, 1769–1780, https://doi.org/10.1093/mp/sst080 (2013).
Ogata, H. et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27, 29–34, https://doi.org/10.1093/nar/27.1.29 (1999).
Li, Y. T. et al. Transcriptome analysis of immune-related genes in Sesarmops sinensis hepatopancreas in reaction to peptidoglycan challenge. Genomics 113, 946–954, https://doi.org/10.1016/j.ygeno.2021.01.011 (2021).
Cock, P. J. et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38, 1767–1771, https://doi.org/10.1093/nar/gkp1137 (2010).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360, https://doi.org/10.1038/nmeth.3317 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295, https://doi.org/10.1038/nbt.3122 (2015).
Li, X. et al. The Manchurian Walnut Genome: Insights into Juglone and Lipid Biosynthesis. Gigascience 11, https://doi.org/10.1093/gigascience/giac057 (2022).
Yu, G. et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287, https://doi.org/10.1089/omi.2011.0118 (2012).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP665145 (2026).
Chen P. Y. et al. MetaboLights MTBLS13314 https://www.ebi.ac.uk/metabolights/MTBLS13314 (2025).
Chen, P. Y. Metabolomic and Transcriptomic Profiling of Two Closely Related Species within the Genus Oldenlandia. figshare https://doi.org/10.6084/m9.figshare.28853480 (2025).
Acknowledgements
This work was supported by the Special Training Plan for Minority Science and Technology Talents, Chinese Medicine of Hubei University of Chinese Medicine (2023ZZXZ002), and Tianshan Innovation Team Program (2020D14030). We gratefully acknowledge Bioyi Biotechnology Co., Ltd., Wuhan, China for providing metabolomics and transcriptomics services.
Author information
Authors and Affiliations
Contributions
Pengyu Chen: Select topic, research proposal design, data analysis and paper writing. Zhuang Huang: Select topic and data analysis. Ping Huang: data analysis. Kaojiang Zhu, Benjiang Xiao and Meng Chen: data sorting. Yuxin Wen and Qi Jiang: Data organization and visualization. Rui Qian, Xing Hong, and Shihao Li: Data review and manuscript review. Fang Huang and Lintao Han: review and editing, experimental funding supporting.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, P., Huang, Z., Wen, Y. et al. Metabolomic and Transcriptomic Profiling of Two Closely Related Species within the Genus Oldenlandia. Sci Data (2026). https://doi.org/10.1038/s41597-026-06745-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06745-y