Abstract
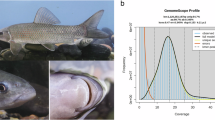
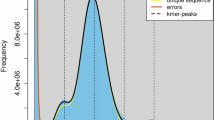
The Siberian crane (Leucogeranus leucogeranus) is classified as Critically Endangered by the IUCN. Its current estimated population is over 6,900 individuals in East Asia, whereas the Western/Central Asian population is nearly extinct, with no recent records of its presence in the wild. Here, we present a high-quality, chromosome-level genome assembly of the Siberian crane generated by integrating Nanopore long-read data, MGISEQ-2000 short-read data, and Hi-C technology data. The assembled genome spans 1.31 Gb, with a scaffold N50 of 83.45 Mb, comprising 33 chromosomes and additional unplaced scaffolds. BUSCO assessment indicated that 97.3 percent of genes in the genome assembly are complete. We identified 10.9 percent repetitive sequences and 21,678 protein-coding genes, of which 88 percent were successfully assigned functional annotations. This high-quality genome assembly and annotation provide a valuable genomic resource for comparative genomic research aimed at understanding the ecology, evolutionary adaptations, and development of Gruidae birds.
Similar content being viewed by others
Data availability
The Hi-C data described in this study are available at in the NCBI Sequence Read Archive database with accession number SRR35316027 (https://www.ncbi.nlm.nih.gov/sra/SRP618574). The sequencing data obtained from the MGISEQ-2000 platforms are deposited into NCBI Sequence Read Archive database with accession number SRR35316036-42 (https://www.ncbi.nlm.nih.gov/sra/SRP618574). The genome assembly is deposited into the DDBJ/ENA/GenBank with accession number JBQWBR000000000 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_053455625.1). The annotation files are available from Figshare (https://doi.org/10.6084/m9.figshare.30017956). All data are publicly available and includes raw sequencing reads, assembled genome, genome annotation files, functional annotation results. Metadata describing the sample information, sequencing platforms, and assembly statistics are also provided in the same repository.
Code availability
The assembly and annotation were performed following the manuals of the corresponding bioinformatics tools with default parameters. The code of the quality assessment and result visualization is available at https://github.com/ChenqCQ/Siberian_crane_Chromosome.
References
BirdLife International. Species factsheet: Leucogeranus leucogeranus. http://www.birdlife.org (2025).
Mirande, C. M. & Harris, J. T. in Crane Conservation Strategy (Baraboo, Wisconsin, USA: International Crane Foundation Press, (2019).
Dussex, N. Comparative Population Genomics Reveal the Determinants of Genome Erosion in Two Sympatric Neotropical Falcons. Mol. Ecol. 34, e17686, https://doi.org/10.1111/mec.17686 (2025).
Theissinger, K. et al. How genomics can help biodiversity conservation. Trends Genet. 39, 545–559, https://doi.org/10.1016/j.tig.2023.01.005 (2023).
Kaewmad, P. et al. First Karyological Analysis of Black the Crowned Crane (Balearica pavonina) and the Scaly-Breasted Munia (Lonchura punctulata). Cytologia 78, 205–211, https://doi.org/10.1508/CYTOLOGIA.78.205 (2013).
Chen, Q. et al. Understanding the Past to Preserve the Future: Genomic Insights Into the Conservation Management of a Critically Endangered Waterbird. Mol. Ecol. 34, e17606, https://doi.org/10.1111/mec.17606 (2025).
Amarasinghe, S. L. et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, https://doi.org/10.1186/s13059-020-1935-5 (2020).
Chen, Y. et al. SOAPnuke: a mapreduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7, 1–6, https://doi.org/10.1093/gigascience/gix120 (2018).
Marcais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770, https://doi.org/10.1093/bioinformatics/btr011 (2011).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204, https://doi.org/10.1093/bioinformatics/btx153 (2017).
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R. & Phillippy, A. M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736, https://doi.org/10.1101/gr.215087.116 (2016).
Vaser, R., Sovic, I., Nagarajan, N. & Sikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746, https://doi.org/10.1101/gr.214270.116 (2017).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963, https://doi.org/10.1371/journal.pone.0112963 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, https://doi.org/10.1093/bioinformatics/btp324 (2009).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259, https://doi.org/10.1186/s13059-015-0831-x (2015).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95, https://doi.org/10.1126/science.aal3327 (2017).
Durand, N. C. et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Systems 3, 99–101, https://doi.org/10.1016/j.cels.2015.07.012 (2016).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212, https://doi.org/10.1093/bioinformatics/btv351 (2015).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 245, https://doi.org/10.1186/s13059-020-02134-9 (2020).
He, W. et al. NGenomeSyn: an easy-to-use and flexible tool for publication-ready visualization of syntenic relationships across multiple genomes. Bioinformatics 39, btad121, https://doi.org/10.1093/bioinformatics/btad121 (2023).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094-3100, https://doi.org/10.1093/bioinformatics/bty191 (2018).
Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. 25, 1–14, https://doi.org/10.1002/0471250953.bi0410s05 (2004).
Ou, S. et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275, https://doi.org/10.1186/s13059-019-1905-y (2019).
Salamov, A. A. & Solovyev, V. V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522, https://doi.org/10.1101/gr.10.4.516 (2000).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644, https://doi.org/10.1093/bioinformatics/btn013 (2008).
Li, H. Protein-to-genome alignment with miniprot. Bioinformatics 39, btad014, https://doi.org/10.1093/bioinformatics/btad014 (2023).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491–504, https://doi.org/10.1186/1471-2105-12-491 (2011).
Zhang, G. et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320, https://doi.org/10.1126/science.1251385 (2014).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 428, 726–731, https://doi.org/10.1016/j.jmb.2015.11.006 (2016).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169, https://doi.org/10.1093/nar/gkw1099 (2016).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410, https://doi.org/10.1006/jmbi.1990.9999 (1990).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360, https://doi.org/10.1093/nar/gky1100 (2018).
The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338, https://doi.org/10.1093/nar/gky1055 (2018).
Chen, Q. Chromosome-level genome assembly and annotation of the critically endangered Siberian crane (Leucogeranus leucogeranus). figshare. Dataset. https://doi.org/10.6084/m9.figshare.30017956.v1 (2025).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 25, 955–964, https://doi.org/10.1093/nar/25.5.955 (1997).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124, https://doi.org/10.1093/nar/gki081 (2005).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935, https://doi.org/10.1093/bioinformatics/btt509 (2013).
Chen, Q. Chromosome-level genome assembly and annotation of the critically endangered Siberian crane (Leucogeranus leucogeranus). NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP618574 (2025).
Chen, Q. Chromosome-level genome assembly and annotation of the critically endangered Siberian crane (Leucogeranus leucogeranus). NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_053455625.1 (2025).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32160132, 32471732) and the Forestry Administration of Guangdong Province, China (DFGP Project of Fauna of Guangdong-202115; Science and Technology Planning Projects of Guangdong Province-2021B1212110002). We appreciate the technical support from the Beijing Genomics Institute (BGI) and EasyATCG Science and Technology Company for sequencing, assembly, and annotation. We appreciate Dr. Russell Doughty for his substantial contributions to improving the readability of this manuscript.
Author information
Authors and Affiliations
Contributions
Yang Liu and Wenjuan Wang conceived and designed the experiments. Peng Huang, Nianhua Dai, and Marria Vladimirtseva collected the samples. Qing Chen performed quality assessment and analyzed the data. Qing Chen wrote the manuscript. Chenqing Zheng, Yang Liu, and Wenjuan Wang reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Q., Zheng, C., Huang, P. et al. Chromosome-level genome assembly and annotation of the critically endangered Siberian crane (Leucogeranus leucogeranus). Sci Data (2026). https://doi.org/10.1038/s41597-026-06773-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06773-8