Figure 1
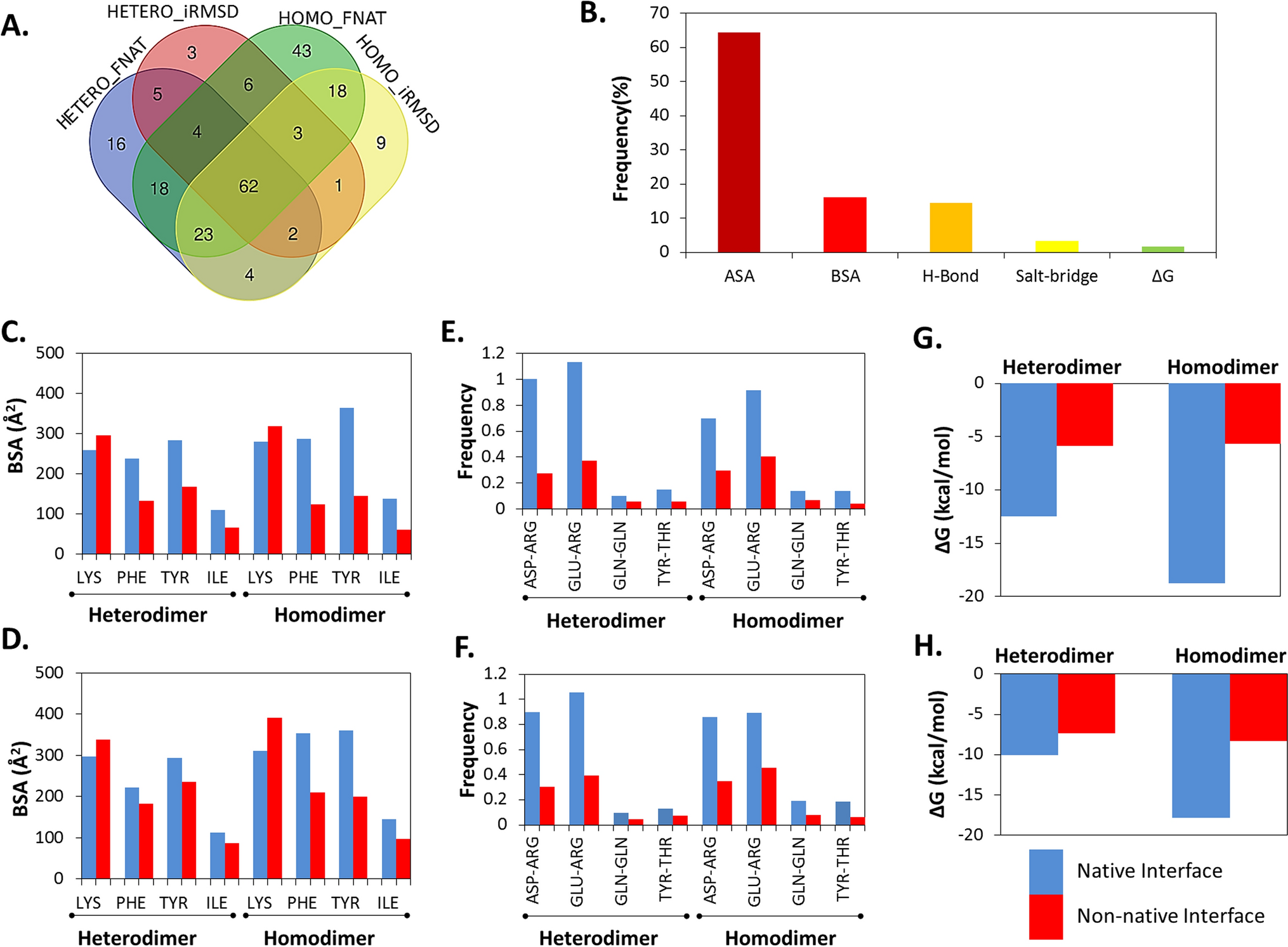
Comparison of protein–protein interaction interface properties. (A) The overlap among interface properties that were showing statistically significant (p ≤ 0.01) differences between the native and non-native like complexes, categorized either by FNAT and iRMSD criteria. HETERO_FNAT and HETERO_iRMSD provide numbers of significantly different interface properties for heterodimers while HOMO_FNAT and HOMO_iRMSD provide numbers of significantly different interface properties for homodimers native and non-native like complexes, respectively. FNAT, fraction of conserved native contacts. iRMSD, interface root mean square deviation. (B) The distribution of the common interface properties that showed statistically significant (p ≤ 0.01) differences between all the native and non-native like complexes. ASA, accessible surface area. BSA, buried surface area. H-bond, hydrogen bonds. (C,D) plot the buried surface area (BSA) of the two amino acids that possessed significantly different BSA at the native interfaces compared to the non-native ones identified based on FNAT (C) and iRMSD (D) definitions, respectively. (E,F) show the hydrogen bond forming amino acid pairs that are found to be significantly higher at the native interfaces compared to the non-native ones identified based on FNAT (E) and iRMSD (F) based definitions, respectively. (G,H) plot the average binding energy represented by ΔG for the native and non-native interfaces identified based on FNAT (G) and iRMSD (H) based definitions, respectively.
