Figure 1
From: Strain-level profiling of viable microbial community by selective single-cell genome sequencing
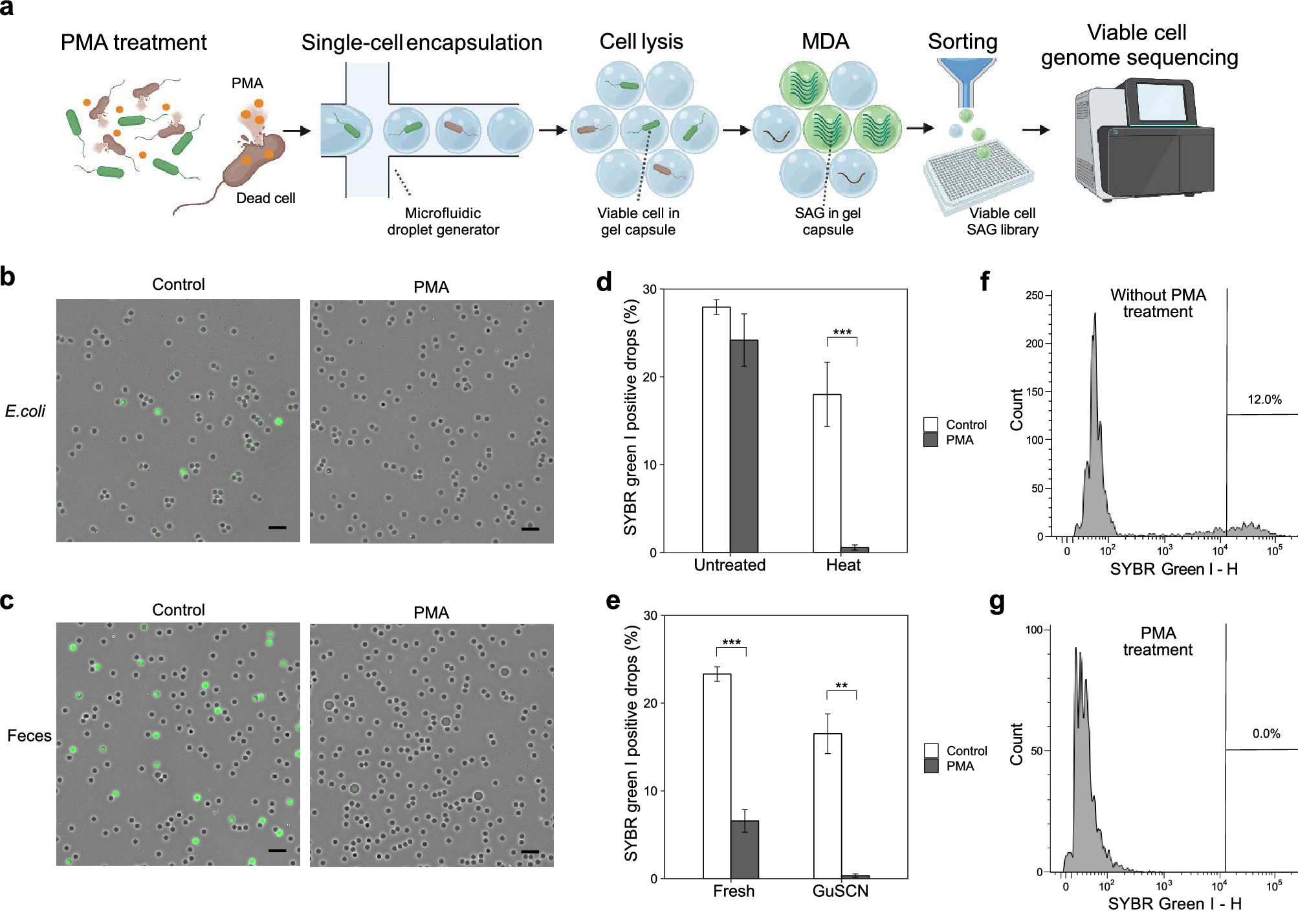
Viable bacteria-targeted single-cell genome amplification with PMA-SAG-gel. (a) Workflow of PMA-SAG-gel for single-cell genome sequencing of viable bacterial cells. The bacterial suspension was treated with PMA to prevent DNA amplification from the dead cells. The cells were then randomly captured in gel capsules via a microfluidic droplet generator and processed via in-gel cell lysis and multiple displacement amplification (MDA) in a test tube. Single-amplified genomes (SAGs) obtained from viable cells were fluorescently detected in the gel and sorted into well plates as SAG library for DNA sequencing. (b, c) Fluorescent images of gel capsules stained with SYBR green I after MDA from heat-killed E. coli (b) and feces suspended in guanidine thiocyanate (GuSCN) solution (c). Green signals indicate amplified genomes in gel capsules. Control indicates no PMA treatment. Scale bar; 100 μm. (d, e) Number of gel capsules stained with SYBR green I after MDA from untreated or heat-killed E. coli (d) and fresh feces or feces suspended in GuSCN solution (e). Data are presented as the mean ± SD (n = 3). **p < 0.01; ***p < 0.005 (Student’s t-test). (f, g) Fluorescent intensity histogram of gel capsules analyzed on a flow cytometer using feces suspended in GuSCN without (f) or with (g) PMA treatment. The gates show the gel capsules to be sorted for subsequent sequencing analysis.
