Figure 1
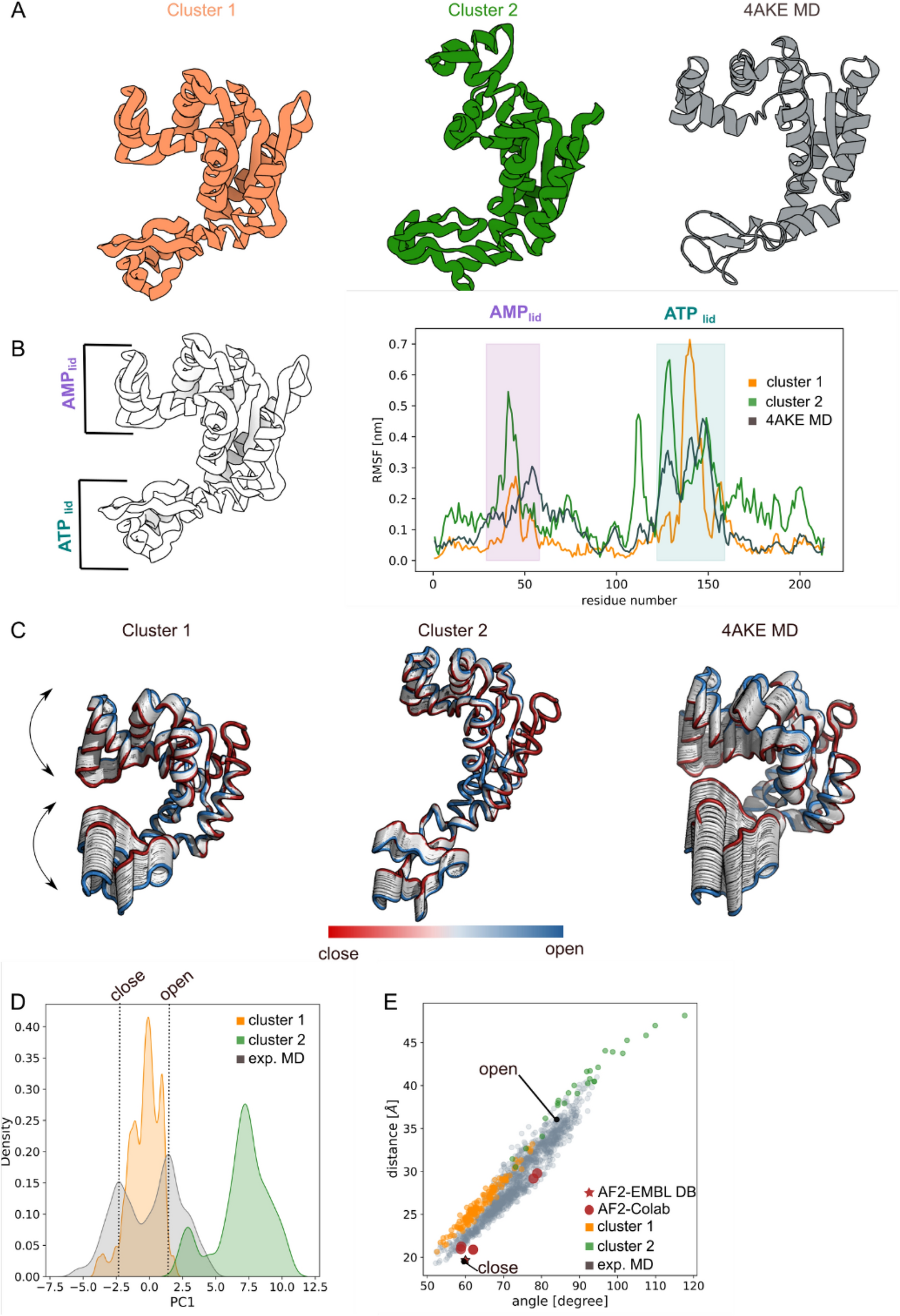
Adenylate kinase. (A) Cartoon representation of the cluster centroids (orange and green) and 4AKE-MD snapshot (grey); (B) Root Mean Square Fluctuation (RMSF) analysis showed a cluster flexibility in the ATPlid and AMPlid regions comparable to the one observed in the 4AKE-MD simulation; (C) Principal Component Analysis for both clusters and MD simulations highlighted the protein internal movement showing for cluster-2 wider conformations than 4AKE-MD simulation; (D) First principal component density distribution plot for 4AKE-MD (grey), cluster-1 (orange) and cluster-2 (green) unveiled that the cluster internal movement explored is compatible with the 4AKE-MD simulation; (E) distance/angle correlation analysis of cluster-1 (orange) and cluster-2 (green) overlap with the conformational space explored by the 4AKE-MD closed and open conformations (gray). AlphaFold2 prediction from EMBLDB (red star) and Google Colab (red spheres) are also reported. The closed and open X-ray adenylate kinase conformation are specified with a black dot. The images were generated using 3D protein imaging webserver (https://3dproteinimaging.com/protein-imager/).
