Figure 1
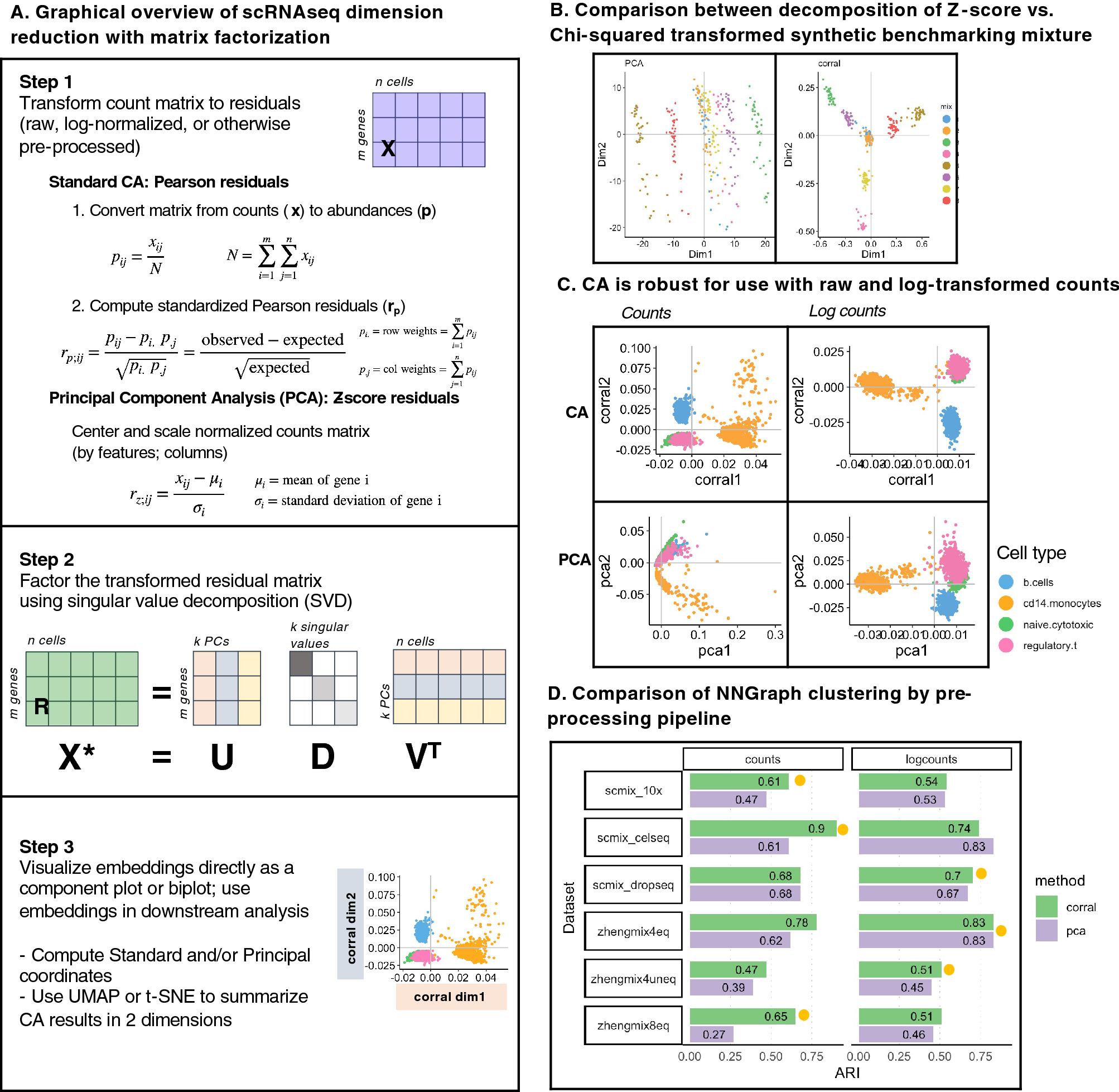
Correspondence Analysis (CA) is an alternative to PCA for count data that is robust for use with raw and log-normalized counts. (A) Graphical overview of steps for dimension reduction with matrix factorization, including standard CA and PCA. Standard CA and PCA can be computed with singular value decomposition (SVD) of the Pearson or the Z-score residuals, respectively. (B) Plots show the first two components generated from PCA (on logcounts; left) and from CA (corral on counts; right) applied to a synthetic benchmarking mRNA mixture with 8 groups (data distributed in the CellBench R package; adapted from3). “Cells” are colored by group. CA resolves the groups into clusters, whereas standard PCA is driven by a gradient in the second component and fails to resolve the groups. (C) Plots show the first two components generated by CA (corral; top row) and PCA (bottom row) on both counts (left column) and logcounts (right column) of the Zhengmix4eq dataset, which comprises approximately 4,000 purified PBMCs in approximately equal mixtures. Cells are colored by type. CA is robust for use with counts or logcounts, whereas PCA on counts results in a horseshoe (arch) effect. (D) CA (green) and PCA (purple) were applied to counts (left column) and logcounts (right column) from six benchmarking datasets (SCMixology; Zhengmix). Embeddings from all approaches were used as input for NNGraph clustering, with performance in recovering published clusters assessed using Adjusted Rand Index (ARI). CA consistently meets or exceeds performance of PCA. Orange circles mark highest ARI achieved in each dataset.
