Abstract
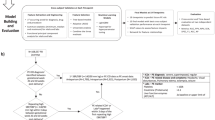
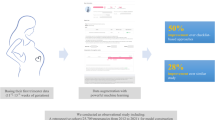
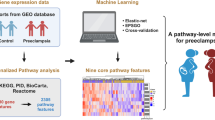
Preeclampsia (PE) is a disease that seriously threatens the health of pregnant women, and early intervention significantly reduce its incidence in high-risk mothers. To identify PE high-risk mother with a certain level of confidence or higher, we aimed to develop a framework for early prediction of preeclampsia (PE), which provides risk scores along with its associated uncertainty score due to missing data in clinical datasets. We built a machine learning model using a multi-center retrospective clinical dataset of 31,235 singleton pregnancies. We assessed the contribution of each variable to prediction variability using Shapley Additive Explanation (SHAP) values in order to quantify uncertainty score resulting from missing data. The score for each sample was calculated by summing the contributions of missing variables. Predictive performance was evaluated using samples with uncertainty scores below specific thresholds, with validation conducted via internal validation and external validation on an independent cohort. Internal validation revealed a strong inverse correlation between uncertainty score thresholds and AUROC (Spearman correlation coefficient: -0.999). At the threshold of 0.11 of the minimum possible level, the AUROC reached 0.978, compared to 0.845 when uncertainty was not considered. In external validation, the AUROC reached 0.994 at the same threshold, compared to 0.693 when uncertainty was not considered. Our framework demonstrated high predictive performance in low-uncertainty samples, emphasizing its stability and effectiveness. This approach reduces the risk of overconfidence in high-uncertainty predictions and represents a more reliable method for PE prediction.
Similar content being viewed by others
Data availability
The datasets generated and/or analysed during the current study are not publicly available due to data protection regulations but are available from the corresponding author on reasonable request.
References
Jeyabalan, A. Epidemiology of preeclampsia: impact of obesity. Nutr. Rev. 71(Suppl 1). https://doi.org/10.1111/nure.12055 (2013). S18-25.
Suksai, M. et al. Preeclampsia and timing of delivery: disease severity, maternal and perinatal outcomes. Pregnancy Hypertens. 37, 101151. https://doi.org/10.1016/j.preghy.2024.101151 (2024).
Sharma, K. J., Alyson, F. E. T., Caughey, A. B. & G., M., B. R. & and Pregnancies complicated by both preeclampsia and growth restriction between 34 and 37 weeks’ gestation are associated with adverse perinatal outcomes. J. Maternal-Fetal Neonatal Med. 30, 2342–2345. https://doi.org/10.1080/14767058.2016.1248394 (2017).
Rolnik, D. L. et al. Aspirin versus placebo in pregnancies at high risk for preterm preeclampsia. New. Engl. J. Med. 377, 613–622. https://doi.org/10.1056/NEJMoa1704559 (2017).
Ansbacher-Feldman, Z. et al. Machine-learning-based prediction of pre-eclampsia using first-trimester maternal characteristics and biomarkers. Ultrasound Obst Gyn. 60, 739–745. https://doi.org/10.1002/uog.26105 (2022).
Tarca, A. L. et al. Prediction of preeclampsia throughout gestation with maternal characteristics and biophysical and biochemical markers: a longitudinal study. Am. J. Obstet. Gynecol. 226 https://doi.org/10.1016/j.ajog.2021.01.020 (2022). 126.e121-126.e122.
Marić, I. et al. Early prediction of preeclampsia via machine learning. Am. J. Obstet. Gynecol. MFM. 2, 100100. https://doi.org/10.1016/j.ajogmf.2020.100100 (2020).
Cohen, J. L. et al. Predictive value of combined serum biomarkers for adverse pregnancy outcomes. Eur. J. Obstet. Gynecol. Reproductive Biology. 181, 89–94 (2014).
Zhou, J., Zhao, X., Wang, Z. & Hu, Y. Combination of lipids and uric acid in mid-second trimester can be used to predict adverse pregnancy outcomes. J. Maternal-Fetal Neonatal Med. 25, 2633–2638. https://doi.org/10.3109/14767058.2012.704447 (2012).
Widmer, M. et al. Accuracy of angiogenic biomarkers at ⩽20weeks’ gestation in predicting the risk of pre-eclampsia: A WHO multicentre study. Pregnancy Hypertension: Int. J. Women’s Cardiovasc. Health. 5, 330–338. https://doi.org/10.1016/j.preghy.2015.09.004 (2015).
Kovacheva, V. P. et al. Preeclampsia prediction using machine learning and polygenic risk scores from clinical and genetic risk factors in early and late pregnancies. Hypertension 81, 264–272. https://doi.org/10.1161/HYPERTENSIONAHA.123.21053 (2024).
Gunderson, E. P. et al. Early pregnancy systolic blood pressure patterns predict Early- and Later‐Onset preeclampsia and gestational hypertension among ostensibly Low‐to‐Moderate risk groups. J. Am. Heart Association. 12, e029617. https://doi.org/10.1161/JAHA.123.029617 (2023).
Lee, S. M. et al. Development of early prediction model for pregnancy-associated hypertension with graph-based semi-supervised learning. Sci. Rep. 12, 15793. https://doi.org/10.1038/s41598-022-15391-4 (2022).
Sandström, A., Snowden, J. M., Bottai, M., Stephansson, O. & Wikström, A. K. Routinely collected antenatal data for longitudinal prediction of preeclampsia in nulliparous women: a population-based study. Sci. Rep. 11, 17973. https://doi.org/10.1038/s41598-021-97465-3 (2021).
Chen, Y. et al. Machine-learning predictive model of pregnancy-induced hypertension in the first trimester. Hypertens. Res. 46, 2135–2144. https://doi.org/10.1038/s41440-023-01298-8 (2023).
Sandström, A., Snowden, J. M., Höijer, J., Bottai, M. & Wikström, A. K. Clinical risk assessment in early pregnancy for preeclampsia in nulliparous women: A population based cohort study. PloS One. 14, e0225716 (2019).
Lahoti, P., Gummadi, K. & Weikum, G. Responsible model deployment via model-agnostic uncertainty learning. Mach. Learn. 112, 939–970. https://doi.org/10.1007/s10994-022-06248-y (2023).
Seoni, S. et al. Application of uncertainty quantification to artificial intelligence in healthcare: A review of last decade (2013–2023). Comput. Biol. Med. 165, 107441. https://doi.org/10.1016/j.compbiomed.2023.107441 (2023).
Martín Vicario, C. et al. Uncertainty-aware deep learning for trustworthy prediction of long-term outcome after endovascular thrombectomy. Sci. Rep. 14, 5544. https://doi.org/10.1038/s41598-024-55761-8 (2024).
Brown, M. A. et al. The hypertensive disorders of pregnancy: ISSHP classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 13, 291–310. https://doi.org/10.1016/j.preghy.2018.05.004 (2018).
Cui, Y., Zhu, B. & Zheng, F. Low-dose aspirin at ≤ 16 weeks of gestation for preventing preeclampsia and its maternal and neonatal adverse outcomes: A systematic review and meta-analysis. Exp. Ther. Med. 15, 4361–4369. https://doi.org/10.3892/etm.2018.5972 (2018).
Skjærven, R., Wilcox, A. J. & Lie, R. T. The interval between pregnancies and the risk of preeclampsia. New. Engl. J. Med. 346, 33–38. https://doi.org/10.1056/NEJMoa011379 (2002).
Stekhoven, D. J. & Bühlmann, P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics 28, 112–118. https://doi.org/10.1093/bioinformatics/btr597 (2011).
Lundberg, S. M. & Lee, S. I. A unified approach to interpreting model predictions. Adv Neur in 30 (2017).
Berisha, V. et al. Digital medicine and the curse of dimensionality. NPJ Digit. Med. 4, 153 (2021).
Acknowledgements
This research was supported by grants through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant numbers: RS-2022-KH129953 and RS-2025-02213591) and by the National Institute of Health (NIH) research project (Project No. 2025-ER1103-01).
Author information
Authors and Affiliations
Contributions
Study concept and design: J.W.K. and N.K.; Data acquisition: Y.J.H., H.J.P., H.Y.B., D.W.K., S.H.J. and E.H.A.; Data preprocessing: J.W.K, H.J.H., J.Y.K, and S.J.Y; Machine learning and Performance evaluation: J.W.K.; Analysis and Interpretation: N.K., J.H.L. and H.M.R.; Drafting of the manuscript: J.W.K and N.K; Revision of the manuscript: N.K., J.H.L. and H.M.R.; Study supervision: J.H.L. and H.M.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, J.W., Kim, N., Kim, J.Y. et al. A novel approach to preeclampsia early prediction addressing predictive uncertainty due to missing data in clinical dataset. Sci Rep (2026). https://doi.org/10.1038/s41598-025-27801-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-27801-4