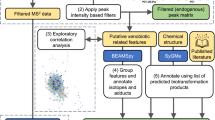
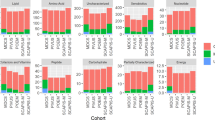
Abstract
Metabolome studies in forensic toxicology focus on the search for endogenous biomarkers changed by, e.g., drugs of abuse. However, placebo-controlled studies, the ideal study design, in humans are scarce for ethical reasons. Thus, the idea of using routine samples became popular, although confounding factors cannot be controlled. To systematically evaluate the use of routine samples for metabolomics, a comparison between a placebo-controlled amphetamine study in humans (A, npos=18, nneg=18) to routine samples either positive or negative for amphetamine, prepared and analyzed over six months (re-evaluated, B, npos=28, nneg=35) and prepared and analyzed within a single analytical batch (re-extracted, C) was performed. Samples were analyzed using untargeted liquid chromatography-tandem-mass-spectrometry. Comparison was conducted on feature level and based on significance (p- and fold-change-values). Only 3 features were significant in A, B, and C, and 2 were identified as amphetamine-(fragments). All 31 significant features from A were present in B and C; however, only 11 (36%) and 4 (13%) of them were significant mainly because of higher variation. Still, other significant features were found in routine samples (B/C).
In conclusion, routine samples are generally suitable for detecting differences in the metabolome, even if they do not match those of a controlled study.
Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to ethical constriction regarding the private information present in routine data. Data can only be made available via the corresponding author upon reasonable request.
References
Xiao, Y. et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell. Res. 32, 477–490. https://doi.org/10.1038/s41422-022-00614-0 (2022).
He, X., Liu, X., Zuo, F., Shi, H. & Jing, J. Artificial intelligence-based multi-omics analysis fuels cancer precision medicine. Semin Cancer Biol. 88, 187–200. https://doi.org/10.1016/j.semcancer.2022.12.009 (2023).
Barberis, E. et al. Precision medicine approaches with metabolomics and artificial intelligence. Int. J. Mol. Sci. 23 https://doi.org/10.3390/ijms231911269 (2022).
Zhang, A., Sun, H., Yan, G., Wang, P. & Wang, X. Mass spectrometry-based metabolomics: applications to biomarker and metabolic pathway research. Biomed. Chromatogr. 30, 7–12. https://doi.org/10.1002/bmc.3453 (2016).
Klein, M. S. & Shearer, J. Metabolomics and type 2 diabetes: Translating basic research into clinical application. J. Diabetes Res. 2016, 3898502. https://doi.org/10.1155/2016/3898502 (2016).
Wang, X., Chen, S. & Jia, W. Metabolomics in cancer biomarker research. Curr. Pharmacol. Rep. 2, 293–298. https://doi.org/10.1007/s40495-016-0074-x (2016).
Ambati, C. S., Yuan, F., Abu-Elheiga, L. A., Zhang, Y. & Shetty, V. Identification and quantitation of malonic acid biomarkers of In-Born error metabolism by targeted metabolomics. J. Am. Soc. Mass. Spectrom. 28, 929–938. https://doi.org/10.1007/s13361-017-1631-1 (2017).
Ren, S. et al. Integration of metabolomics and transcriptomics reveals major metabolic pathways and potential biomarker involved in prostate cancer. Mol. Cell. Proteom. 15, 154–163. https://doi.org/10.1074/mcp.M115.052381 (2016).
Wurtz, P. et al. Metabolic profiling of alcohol consumption in 9778 young adults. Int. J. Epidemiol. 45, 1493–1506. https://doi.org/10.1093/ije/dyw175 (2016).
Lu, Y. & Chen, C. Metabolomics: Bridging chemistry and biology in drug discovery and development. Curr. Pharmacol. Rep. 3, 16–25. https://doi.org/10.1007/s40495-017-0083-4 (2017).
Mercier, K. A., Al-Jazrawe, M., Poon, R., Acuff, Z. & Alman, B. A metabolomics pilot study on desmoid tumors and novel drug candidates. Sci. Rep. 8, 584. https://doi.org/10.1038/s41598-017-18921-7 (2018).
Castillo-Peinado, L. S. & de Luque, M. D. Present and foreseeable future of metabolomics in forensic analysis. Anal. Chim. Acta. 925, 1–15. https://doi.org/10.1016/j.aca.2016.04.040 (2016).
Steuer, A. E., Brockbals, L. & Kraemer, T. Metabolomic strategies in biomarker Research-New approach for indirect identification of drug consumption and sample manipulation in clinical and forensic toxicology? Front. Chem. 7, 319. https://doi.org/10.3389/fchem.2019.00319 (2019).
Steuer, A. E., Brockbals, L. & Kraemer, T. Untargeted metabolomics approaches to improve casework in clinical and forensic toxicology-Where are we standing and where are we heading? Wires Forensic Sci. 4, e1449 (2021).
Manier, S. K. & Meyer, M. R. Current situation of the metabolomics techniques used for the metabolism studies of new psychoactive substances. Ther. Drug Monit. 42, 93–97. https://doi.org/10.1097/FTD.0000000000000694 (2020).
Szeremeta, M., Pietrowska, K., Niemcunowicz-Janica, A., Kretowski, A. & Ciborowski, M. Applications of metabolomics in forensic toxicology and forensic medicine. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms22063010 (2021).
Dinis-Oliveira, R. J. Metabolomics of drugs of abuse: a more realistic view of the toxicological complexity. Bioanalysis 6, 3155–3159. https://doi.org/10.4155/bio.14.260 (2014).
Zaitsu, K., Hayashi, Y., Kusano, M., Tsuchihashi, H. & Ishii, A. Application of metabolomics to toxicology of drugs of abuse: A mini review of metabolomics approach to acute and chronic toxicity studies. Drug Metab. Pharmacokinet. 31, 21–26. https://doi.org/10.1016/j.dmpk.2015.10.002 (2016).
Nielsen, K. L., Telving, R., Andreasen, M. F., Hasselstrom, J. B. & Johannsen, M. A. A metabolomics study of retrospective forensic data from whole blood samples of humans exposed to 3,4-methylenedioxymethamphetamine: A new approach for identifying drug metabolites and changes in metabolism related to drug consumption. J. Proteome Res. 15, 619–627. https://doi.org/10.1021/acs.jproteome.5b01023 (2016).
Wang, T. et al. A retrospective metabolomics analysis of Gamma-Hydroxybutyrate in humans: new potential markers and changes in metabolism related to GHB consumption. Front. Pharmacol. 13, 816376. https://doi.org/10.3389/fphar.2022.816376 (2022).
Pasin, D. et al. Metabolomics-driven determination of targets for Salicylic acid and ibuprofen in positive electrospray ionization using LC-HRMS. Drug Test. Anal. 14, 747–756. https://doi.org/10.1002/dta.3215 (2022).
Mollerup, C. B. et al. Retrospective analysis for valproate screening targets with liquid chromatography-high resolution mass spectrometry with positive electrospray ionization: an omics-based approach. Drug Test. Anal. 11, 730–738. https://doi.org/10.1002/dta.2543 (2019).
Hoj, L. J. et al. Identification of phenobarbital and other barbiturates in forensic drug screening using positive electrospray ionization liquid chromatography-high resolution mass spectrometry. Drug Test. Anal. 11, 1258–1263. https://doi.org/10.1002/dta.2603 (2019).
Ward, L. J. et al. Postmortem metabolomics as a high-throughput cause-of-death screening tool for human death investigations. iScience 27, 109794. https://doi.org/10.1016/j.isci.2024.109794 (2024).
Kronstrand, R. et al. The metabolism of the synthetic cannabinoids ADB-BUTINACA and ADB-4en-PINACA and their detection in forensic toxicology casework and infused papers seized in prisons. Drug Test. Anal. 14, 634–652. https://doi.org/10.1002/dta.3203 (2022).
Roman, M., Strom, L., Tell, H. & Josefsson, M. Liquid chromatography/time-of-flight mass spectrometry analysis of postmortem blood samples for targeted toxicological screening. Anal. Bioanal Chem. 405, 4107–4125. https://doi.org/10.1007/s00216-013-6798-0 (2013).
Brockbals, L. et al. Time- and Site-Dependent postmortem redistribution of antidepressants and neuroleptics in blood and alternative matrices. J. Anal. Toxicol. 45, 356–367. https://doi.org/10.1093/jat/bkaa092 (2020).
Brockbals, L. et al. Postmortem metabolomics: strategies to assess time-dependent postmortem changes of diazepam, nordiazepam, morphine, codeine, mirtazapine and citalopram. Metabolites 11, 643 (2021).
Steuer, A. E. et al. Identification of new urinary gamma-hydroxybutyric acid markers applying untargeted metabolomics analysis following placebo-controlled administration to humans. Drug. Test. Anal. 11, 813–823. https://doi.org/10.1002/dta.2558 (2019).
Steuer, A. E., Keller, M., Kraemer, T. & Poetzsch, S. N. Multianalyte approach-including automated preparation of calibrators-for validated quantification of 82 drugs in whole blood by liquid Chromatography-Tandem mass spectrometry. Drug Test. Anal. https://doi.org/10.1002/dta.3794 (2024).
Holze, F. et al. Distinct acute effects of LSD, MDMA, and D-amphetamine in healthy subjects. Neuropsychopharmacology 45, 462–471. https://doi.org/10.1038/s41386-019-0569-3 (2020).
Guo, J., Shen, S. & Huan, T. Paramounter: Direct measurement of universal parameters to process metabolomics data in a white box. Anal. Chem. 94, 4260–4268. https://doi.org/10.1021/acs.analchem.1c04758 (2022).
Tsugawa, H. et al. MS-DIAL: data-independent MS/MS Deconvolution for comprehensive metabolome analysis. Nat. Methods. 12, 523–526. https://doi.org/10.1038/nmeth.3393 (2015).
R: A language and environment for statistical computing v. 4.4.1. (R Foundation for Statistical Computing, 2024).
Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for Dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290. https://doi.org/10.1021/ac051632c (2006).
Pang, Z. et al. Using metaboanalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761. https://doi.org/10.1038/s41596-022-00710-w (2022).
Shapiro, S. S. An analysis of variance test for normality (Complete Samples). Biometrika 52, 591–611 (1965).
Steuer, A. E. et al. Comparative untargeted metabolomics analysis of the psychostimulants 3,4-Methylenedioxy-Methamphetamine (MDMA), Amphetamine, and the novel psychoactive substance mephedrone after controlled drug administration to humans. Metabolites 10 https://doi.org/10.3390/metabo10080306 (2020).
Fernandez-Albert, F. et al. Intensity drift removal in LC/MS metabolomics by common variance compensation. Bioinformatics 30, 2899–2905. https://doi.org/10.1093/bioinformatics/btu423 (2014).
Yu, Z. et al. Differences between human plasma and serum metabolite profiles. PLoS One. 6, e21230. https://doi.org/10.1371/journal.pone.0021230 (2011).
Handley, S. A., Silk, S. W., Fisher, D. S., Subramaniam, K. & Flanagan, R. J. Clozapine and Norclozapine concentrations in paired human plasma and serum samples. Ther. Drug Monit. 40, 148–150. https://doi.org/10.1097/ftd.0000000000000478 (2018).
Hirayama, A. et al. Effects of processing and storage conditions on charged metabolomic profiles in blood. Electrophoresis 36, 2148–2155. https://doi.org/10.1002/elps.201400600 (2015).
Chen, D., Han, W., Huan, T., Li, L. & Li, L. Effects of Freeze-Thaw cycles of blood samples on High-Coverage quantitative metabolomics. Anal. Chem. 92, 9265–9272. https://doi.org/10.1021/acs.analchem.0c01610 (2020).
Faul, F., Erdfelder, E., Lang, A. G. & Buchner, A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 39, 175–191. https://doi.org/10.3758/bf03193146 (2007).
Stanislava Rakusanova, T. C. Tips and tricks for LC–MS-based metabolomics and lipidomics analysis. TRAC Trends Anal. Chem. 180 https://doi.org/10.1016/j.trac.2024.117940 (2024). https://doi.org/https://doi.org/
Blaise, B. J. et al. Power analysis and sample size determination in metabolic phenotyping. Anal. Chem. 88, 5179–5188. https://doi.org/10.1021/acs.analchem.6b00188 (2016).
Acknowledgements
The authors would like to thank Maja Keller for her support and express their gratitude to Emma Louise Kessler, MD for her generous legacy she donated to the Institute of Forensic Medicine at the University of Zurich, Switzerland for research purposes.
Author information
Authors and Affiliations
Contributions
Annina Bovens conducted the experiments, the statistical analysis and wrote the manuscript. Claudio Leu conducted the experiments. Lana Brockbals wrote the manuscript. Friederike Holze and Matthias E. Liechti provided the samples from the controlled administration study. Thomas Kraemer wrote the manuscript. Andrea E. Steuer had the organizational lead and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent was obtained for all data used from the placebo-controlled study (study A) performed by Holze et al. The study was registered at ClinicalTrials.gov (NCT03019822) and was in full accordance with the Declaration of Helsinki, as well as approved by the Ethics Committee Northwest Switzerland (EKNZ). Due to the retrospective nature of the routine data used for studies B and C a waiver and a declaration of no objection for ethical approval of the Cantonal Ethics Board of the Canton of Zurich were obtained (KEK waiver no. 42.2005 and BASEC-Nr. Req2017-00946).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bovens, A., Leu, C., Brockbals, L. et al. Comparison of human metabolome changes identified in a placebo-controlled amphetamine administration study versus those using forensic toxicology routine data. Sci Rep (2026). https://doi.org/10.1038/s41598-026-34985-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-34985-w