Abstract
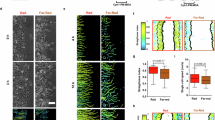
Activation of transmembrane CD13 enables diverse cellular processes such as cell–cell and cell–matrix adhesion, endocytosis, and recycling of cell surface proteins by assembling and tethering protein complexes at the plasma membrane. Here, we identify a novel CD13-dependent protein assembly that regulates phosphoinositide (PI) signal transduction to impact actin dynamics and induce cell protrusions capable of propagating signals to distant cells. In response to cellular stress, the CD13-expressing human Kaposi’s sarcoma-derived cell line (KS1767, KSCs) formed elongated protrusions that extend above the substrate and link non-adjacent cells, which is significantly diminished in CD13KO KSCs. Activation of CD13 with stimulating mAbs markedly induced protrusion formation with a striking accumulation of CD13 and actin at the base. Further, these membrane-delimited bridges in WT KSCs can transfer Ca2+ signals between connected cells via Connexin 43+ gap junctions. Mechanistically, CD13-mediated protrusion formation requires activation of CD13, Src, FAK and Cdc42 to tether the IQGAP1 and ARF6 complex at the membrane. This activates phosphatidylinositol-4-phosphate-5-kinase (PI5K) to increase local phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) levels, promoting actin-polymerization and membrane protrusion. Therefore, CD13 is a novel molecular PIP regulator, modulating signal transduction and downstream cellular processes, including actin cytoskeleton dynamics and membrane organization to facilitate intercellular communication.
Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available from the corresponding authors upon reasonable request.
References
Bezanilla, M., Gladfelter, A. S., Kovar, D. R. & Lee, W. L. Cytoskeletal dynamics: A view from the membrane. J. Cell Biol. 209, 329–337. https://doi.org/10.1083/jcb.201502062 (2015).
Belian, S., Korenkova, O. & Zurzolo, C. Actin-based protrusions at a glance. J. Cell Sci. https://doi.org/10.1242/jcs.261156 (2023).
Yamashita, Y. M., Inaba, M. & Buszczak, M. specialized intercellular communications via cytonemes and nanotubes. Annu. Rev. Cell Dev. Biol. 34, 59–84. https://doi.org/10.1146/annurev-cellbio-100617-062932 (2018).
Delage, E. et al. Differential identity of filopodia and tunneling nanotubes revealed by the opposite functions of actin regulatory complexes. Sci. Rep. 6, 39632. https://doi.org/10.1038/srep39632 (2016).
Schiller, C. et al. LST1 promotes the assembly of a molecular machinery responsible for tunneling nanotube formation. J. Cell Sci. 126, 767–777. https://doi.org/10.1242/jcs.114033 (2013).
D’Aloia, A. et al. RalGPS2 interacts with Akt and PDK1 promoting tunneling nanotubes formation in bladder cancer and kidney cells microenvironment. Cancers https://doi.org/10.3390/cancers13246330 (2021).
Grabowska, W. et al. IQGAP1-dysfunction leads to induction of senescence in human vascular smooth muscle cells. Mech. Ageing Dev. 190, 111295. https://doi.org/10.1016/j.mad.2020.111295 (2020).
Hase, K. et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell. Biol. 11, 1427–1432. https://doi.org/10.1038/ncb1990 (2009).
Barutta, F. et al. Protective role of the M-Sec-tunneling nanotube system in podocytes. J. Am. Soc. Nephrol. 32, 1114–1130. https://doi.org/10.1681/ASN.2020071076 (2021).
Wenk, M. R. The emerging field of lipidomics. Nat. Rev. Drug Discovery 4, 594–610. https://doi.org/10.1038/nrd1776 (2005).
Simons, K. & Sampaio, J. L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697. https://doi.org/10.1101/cshperspect.a004697 (2011).
Delage, E. & Zurzolo, C. Exploring the role of lipids in intercellular conduits: Breakthroughs in the pipeline. Front. Plant Sci. 4, 504. https://doi.org/10.3389/fpls.2013.00504 (2013).
Takenawa, T. & Itoh, T. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim Biophys Acta 1533, 190–206. https://doi.org/10.1016/s1388-1981(01)00165-2 (2001).
Regen, S. L. The origin of lipid rafts. Biochemistry 59, 4617–4621. https://doi.org/10.1021/acs.biochem.0c00851 (2020).
Chichili, G. R. & Rodgers, W. Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci. CMLS 66, 2319–2328. https://doi.org/10.1007/s00018-009-0022-6 (2009).
Saarikangas, J., Zhao, H. & Lappalainen, P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90, 259–289. https://doi.org/10.1152/physrev.00036.2009 (2010).
Caviglia, S. & Ober, E. A. Non-conventional protrusions: The diversity of cell interactions at short and long distance. Curr. Opin. Cell. Biol. 54, 106–113. https://doi.org/10.1016/j.ceb.2018.05.013 (2018).
Naphade, S. et al. Brief reports: Lysosomal cross-correction by hematopoietic stem cell-derived macrophages via tunneling nanotubes. Stem Cells 33, 301–309. https://doi.org/10.1002/stem.1835 (2015).
Rahman, M. M. et al. CD13 regulates anchorage and differentiation of the skeletal muscle satellite stem cell population in ischemic injury. Stem Cells 32, 1564–1577. https://doi.org/10.1002/stem.1610 (2014).
Rahman, M. M. et al. CD13 promotes mesenchymal stem cell-mediated regeneration of ischemic muscle. Front. Physiol. 4, 402. https://doi.org/10.3389/fphys.2013.00402 (2014).
Bhagwat, S. V. et al. CD13/APN is activated by angiogenic signals and is essential for capillary tube formation. Blood 97, 652–659 (2001).
Pasqualini, R. et al. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 60, 722–727 (2000).
Petrovic, N. et al. CD13/APN regulates endothelial invasion and filopodia formation. Blood 110, 142–150. https://doi.org/10.1182/blood-2006-02-002931 (2007).
Ghosh, M., Subramani, J., Rahman, M. M. & Shapiro, L. H. CD13 restricts TLR4 endocytic signal transduction in inflammation. J. Immunol. 194, 4466–4476. https://doi.org/10.4049/jimmunol.1403133 (2015).
Ghosh, M. et al. Molecular mechanisms regulating CD13-mediated adhesion. Immunology 142, 636–647. https://doi.org/10.1111/imm.12279 (2014).
Pereira, F. E. et al. CD13 is essential for inflammatory trafficking and infarct healing following permanent coronary artery occlusion in mice. Cardiovasc. Res. 100, 74–83. https://doi.org/10.1093/cvr/cvt155[pii] (2013).
Mina-Osorio, P., Shapiro, L. H. & Ortega, E. CD13 in cell adhesion: aminopeptidase N (CD13) mediates homotypic aggregation of monocytic cells. J. Leukoc. Biol. 79, 719–730 (2006).
Subramani, J. et al. Tyrosine phosphorylation of CD13 regulates inflammatory cell-cell adhesion and monocyte trafficking. J. Immunol. 191, 3905–3912. https://doi.org/10.4049/jimmunol.1301348 (2013).
Ghosh, M. et al. CD13 tethers the IQGAP1-ARF6-EFA6 complex to the plasma membrane to promote ARF6 activation, beta1 integrin recycling, and cell migration. Sci. Signal https://doi.org/10.1126/scisignal.aav5938 (2019).
Ghosh, M., Kelava, T., Madunic, I. V., Kalajzic, I. & Shapiro, L. H. CD13 is a critical regulator of cell-cell fusion in osteoclastogenesis. Sci. Rep. 11, 10736. https://doi.org/10.1038/s41598-021-90271-x (2021).
Ghosh, M. & Shapiro, L. H. CD13 regulation of membrane recycling: Implications for cancer dissemination. Mol. Cell. Oncol. https://doi.org/10.1080/23723556.2019.1648024 (2019).
Mina-Osorio, P. et al. CD13 is a novel mediator of monocytic/endothelial cell adhesion. J Leukoc Biol 84, 448–459 (2008).
Dubois, F. et al. A role for RASSF1A in tunneling nanotube formation between cells through GEFH1/Rab11 pathway control. Cell Commun. Signal 16, 66. https://doi.org/10.1186/s12964-018-0276-4 (2018).
Desir, S. et al. Chemotherapy-induced tunneling nanotubes mediate intercellular drug efflux in pancreatic cancer. Sci. Rep. 8, 9484. https://doi.org/10.1038/s41598-018-27649-x (2018).
Resnik, N. et al. Molecular, morphological and functional properties of tunnelling nanotubes between normal and cancer urothelial cells: New insights from the in vitro model mimicking the situation after surgical removal of the urothelial tumor. Front. Cell Dev. Biol. 10, 934684. https://doi.org/10.3389/fcell.2022.934684 (2022).
Florey, O., Durgan, J. & Muller, W. Phosphorylation of leukocyte PECAM and its association with detergent-resistant membranes regulate transendothelial migration. J. Immunol. 185, 1878–1886. https://doi.org/10.4049/jimmunol.1001305 (2010).
Liu, G. et al. ICAM-1-activated Src and eNOS signaling increase endothelial cell surface PECAM-1 adhesivity and neutrophil transmigration. Blood 120, 1942–1952. https://doi.org/10.1182/blood-2011-12-397430 (2012).
Berman, M. E. & Muller, W. A. Ligation of platelet/endothelial cell adhesion molecule 1 (PECAM-1/CD31) on monocytes and neutrophils increases binding capacity of leukocyte CR3 (CD11b/CD18). J. Immunol. 154, 299–307 (1995).
Ghosh, M., McAuliffe, B., Subramani, J., Basu, S. & Shapiro, L. H. CD13 regulates dendritic cell cross-presentation and T cell responses by inhibiting receptor-mediated antigen uptake. J. Immunol. 188, 5489–5499. https://doi.org/10.4049/jimmunol.1103490 (2012).
Saenz-de-Santa-Maria, I. et al. Control of long-distance cell-to-cell communication and autophagosome transfer in squamous cell carcinoma via tunneling nanotubes. Oncotarget 8, 20939–20960. https://doi.org/10.18632/oncotarget.15467 (2017).
Tishchenko, A. et al. Cx43 and associated cell signaling pathways regulate tunneling nanotubes in breast cancer cells. Cancers https://doi.org/10.3390/cancers12102798 (2020).
Bhat, S. et al. Rab35 and its effectors promote formation of tunneling nanotubes in neuronal cells. Sci. Rep. 10, 16803. https://doi.org/10.1038/s41598-020-74013-z (2020).
Rohatgi, R. et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231. https://doi.org/10.1016/s0092-8674(00)80732-1 (1999).
Sugihara, K. et al. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol. 4, 73–78. https://doi.org/10.1038/ncb720 (2002).
Hanna, S. J. et al. The role of Rho-GTPases and actin polymerization during macrophage tunneling nanotube biogenesis. Sci. Rep. 7, 8547. https://doi.org/10.1038/s41598-017-08950-7 (2017).
Kimura, S. et al. Distinct roles for the N- and C-terminal regions of M-Sec in plasma membrane deformation during tunneling nanotube formation. Sci. Rep. 6, 33548. https://doi.org/10.1038/srep33548 (2016).
Mandal, K. Review of PIP2 in cellular signaling, functions and diseases. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21218342 (2020).
Brown, F. D., Rozelle, A. L., Yin, H. L., Balla, T. & Donaldson, J. G. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell. Biol. 154, 1007–1017. https://doi.org/10.1083/jcb.200103107 (2001).
Varnai, P. & Balla, T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell. Biol. 143, 501–510. https://doi.org/10.1083/jcb.143.2.501 (1998).
Holz, R. W. et al. A pleckstrin homology domain specific for phosphatidylinositol 4,5-bisphosphate (PtdIns-4,5–P2) and fused to green fluorescent protein identifies plasma membrane PtdIns-4,5–P2 as being important in exocytosis. J. Biol. Chem. 275, 17878–17885. https://doi.org/10.1074/jbc.M000925200 (2000).
Wang, X. & Gerdes, H. H. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ 22, 1181–1191. https://doi.org/10.1038/cdd.2014.211 (2015).
Alarcon-Martinez, L. et al. Interpericyte tunnelling nanotubes regulate neurovascular coupling. Nature 585, 91–95. https://doi.org/10.1038/s41586-020-2589-x (2020).
Lock, J. T., Parker, I. & Smith, I. F. Communication of Ca(2+) signals via tunneling membrane nanotubes is mediated by transmission of inositol trisphosphate through gap junctions. Cell Calcium 60, 266–272. https://doi.org/10.1016/j.ceca.2016.06.004 (2016).
Smith, I. F., Shuai, J. & Parker, I. Active generation and propagation of Ca2+ signals within tunneling membrane nanotubes. Biophys. J . 100, L37-39. https://doi.org/10.1016/j.bpj.2011.03.007 (2011).
Wang, X., Veruki, M. L., Bukoreshtliev, N. V., Hartveit, E. & Gerdes, H. H. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc. Natl. Acad. Sci. U S A 107, 17194–17199. https://doi.org/10.1073/pnas.1006785107 (2010).
Watkins, S. C. & Salter, R. D. Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity 23, 309–318. https://doi.org/10.1016/j.immuni.2005.08.009 (2005).
Onfelt, B., Nedvetzki, S., Yanagi, K. & Davis, D. M. Cutting edge: Membrane nanotubes connect immune cells. J. Immunol. 173, 1511–1513 (2004).
Johnson, L. A. et al. Dendritic cells enter lymph vessels by hyaluronan-mediated docking to the endothelial receptor LYVE-1. Nat. Immunol. 18, 762–770. https://doi.org/10.1038/ni.3750 (2017).
Vestweber, D., Zeuschner, D., Rottner, K. & Schnoor, M. Cortactin regulates the activity of small GTPases and ICAM-1 clustering in endothelium: Implications for the formation of docking structures. Tissue Barriers 1, e23862. https://doi.org/10.4161/tisb.23862 (2013).
Wittchen, E. S. Endothelial signaling in paracellular and transcellular leukocyte transmigration. Front. Biosci. (Landmark edition) 14, 2522–2545 (2009).
Pergu, R. et al. The chaperone ERp29 is required for tunneling nanotube formation by stabilizing MSec. J. Biol. Chem. 294, 7177–7193. https://doi.org/10.1074/jbc.RA118.005659 (2019).
Zhu, S. et al. Rab11a-Rab8a cascade regulates the formation of tunneling nanotubes through vesicle recycling. J. Cell Sci. https://doi.org/10.1242/jcs.215889 (2018).
Gousset, K., Marzo, L., Commere, P. H. & Zurzolo, C. Myo10 is a key regulator of TNT formation in neuronal cells. J. Cell Sci. 126, 4424–4435. https://doi.org/10.1242/jcs.129239 (2013).
Reichert, D. et al. Tunneling nanotubes mediate the transfer of stem cell marker CD133 between hematopoietic progenitor cells. Exp. Hematol. 44, 1092–1112. https://doi.org/10.1016/j.exphem.2016.07.006 (2016).
Pawar, A. et al. Ral-Arf6 crosstalk regulates Ral dependent exocyst trafficking and anchorage independent growth signalling. Cell Signal 28, 1225–1236. https://doi.org/10.1016/j.cellsig.2016.05.023 (2016).
Ghosh, M. et al. The implant-induced foreign body response is limited by CD13-dependent regulation of ubiquitination of fusogenic proteins. J. Immunol. 212, 663–676. https://doi.org/10.4049/jimmunol.2300688 (2024).
Carnicer-Lombarte, A., Chen, S. T., Malliaras, G. G. & Barone, D. G. Foreign body reaction to implanted biomaterials and its impact in nerve neuroprosthetics. Front. Bioeng. Biotechnol. 9, 622524. https://doi.org/10.3389/fbioe.2021.622524 (2021).
Ellerby, H. M. et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 5, 1032–1038 (1999).
Zong, P. et al. Functional coupling of TRPM2 and extrasynaptic NMDARs exacerbates excitotoxicity in ischemic brain injury. Neuron 110, 1944–1958. https://doi.org/10.1016/j.neuron.2022.03.021 (2022).
Acknowledgements
We thank Chris Stoddard at Human Genome Editing Core at UConn Health for the generation of CRISPR cell lines. We also thank Susan Staurovsky at Cell Analysis and Modeling Core at UConn Health for technical help.
Funding
This work was supported by National Institutes of Health grant R21AI15 and Research Excellence grant, University of Connecticut to LHS and MG.
Author information
Authors and Affiliations
Contributions
EM, PZ, LY, LHS and MG designed the research plan. EM, BA, RS, NT, FM, PZ and MG conducted experiments and analyzed data. EM, LHS and MG wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Meredith, E., Aguilera, B., Sharma, R. et al. CD13 activation assembles phosphoinositide (PI) signaling complexes to regulate the actin cytoskeleton. Sci Rep (2026). https://doi.org/10.1038/s41598-026-35022-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-35022-6