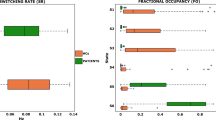
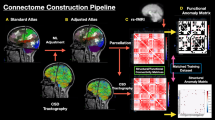
Abstract
Brain tumors, particularly meningiomas and gliomas, can profoundly affect neural function, yet their impact on brain dynamics remains incompletely understood. This study investigates alterations in normal brain function among meningioma and glioma patients by assessing dynamical complexity through the Intrinsic Ignition Framework. We analyzed resting-state fMRI data from 34 participants to quantify brain dynamics using intrinsic ignition and metastability metrics. Our results revealed distinct patterns of disruption: glioma patients showed significant reductions in both metrics compared to controls, indicating widespread network disturbances. In contrast, meningioma patients exhibited significant changes predominantly in regions with substantial tumor involvement. Resting-state network analysis demonstrated strong metastability and metastability/ignition correlations between regions in controls, which were slightly weakened in meningioma patients and severely disrupted in glioma patients. These findings highlight the differential impacts of gliomas and meningiomas on brain function, offering insights into their distinct pathophysiological mechanisms. Furthermore, these results show that brain dynamics metrics can be effective biomarkers for identifying disruptions in brain information transmission caused by tumors.
Similar content being viewed by others
Data availability
This study utilized the Brain Tumor Connectomics Data, which contains pre-operative data from 11 glioma patients, 14 meningioma patients, and 11 control subjects previously described in [35, 38]. The dataset is publicly available at https://doi.org/10.18112/openneuro.ds001226.v5.0.0.
Code availability
All code for implementing and reproducing our results is available at https://github.com/ajunca/BrainTumor.
References
Wiemels, J., Wrensch, M. & Claus, E. B. Epidemiology and etiology of meningioma. J. Neuro oncol.99(3), 307–314. https://doi.org/10.1007/s11060-010-0386-3 (2010).
Louis, David N. et al. The 2016 World Health Organization Classification of tumors of the central nervous system: A summary. Acta Neuropathol.131(6), 803–820. https://doi.org/10.1007/s00401-016-1545-1 (2016).
Ostrom, Q.T., et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. In: Neuro-Oncology 22 (Supplement_1 Oct 30, 2020), pp iv1–iv96. ISSN: 1522-8517, 1523-5866. https://doi.org/10.1093/neuonc/noaa200.
Zong, H., Parada, L. F. & Baker, S. J. Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harb. Perspect. Biol.7(5), a020610. https://doi.org/10.1101/cshperspect.a020610 (2015).
Bou Zerdan, M., et al. Latest updates on cellular and molecular biomarkers of gliomas. Front. Oncol. 12, 1030366. https://doi.org/10.3389/fonc.2022.1030366.
Ogasawara, C., Philbrick, B. D. & Adamson, D. C. Meningioma: A review of epidemiology, pathology, diagnosis, treatment, and future directions. Biomedicines9(3), 319. https://doi.org/10.3390/biomedicines9030319 (2021).
Chieffo, D. P. R. et al. Brain tumor at diagnosis: From cognition and behavior to quality of life. Diagnostics13(3), 541. https://doi.org/10.3390/diagnostics13030541 (2023).
Sporns, O., Tononi, G. & Kötter, R. The Human Connectome: A Structural Description of the Human Brain. In: PLoS Computational Biology 1.4 (2005), e42. ISSN: 1553-734X, 1553-7358. https://doi.org/10.1371/journal.pcbi.0010042.
Fox, M. D. & Raichle, M. E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci.8(9), 700–711. https://doi.org/10.1038/nrn2201 (2007).
Logothetis, N. K. What we can do and what we cannot do with fMRI. Nature453(7197), 869–878. https://doi.org/10.1038/nature06976 (2008).
Deco, G., Jirsa, V. K. & McIntosh, A. R. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat. Rev. Neurosci.12(1), 43–56. https://doi.org/10.1038/nrn2961 (2011).
Stam, C. J. & Reijneveld, J. C. Graph theoretical analysis of complex networks in the brain. Nonlinear Biomed. Phys.1(1), 3. https://doi.org/10.1186/1753-4631-1-3 (2007).
Whitfield-Gabrieli, S. et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. In: Proceedings of the National Academy of Sciences 106.4 (Jan 27, 2009), 1279–1284. ISSN: 0027-8424, 1091-6490. https://doi.org/10.1073/pnas.0809141106.
Englot, D. J. et al. Global and regional functional connectivity maps of neural oscillations in focal epilepsy. Brain138(8), 2249–2262. https://doi.org/10.1093/brain/awv130 (2015).
Deco, G. & Kringelbach, M. L. Great expectations: Using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron84(5), 892–905. https://doi.org/10.1016/j.neuron.2014.08.034 (2014).
Bassett, D. S. & Sporns, O. Network neuroscience. Nat. Neurosci.20(3), 353–364. https://doi.org/10.1038/nn.4502 (2017).
Aerts, H. et al. Brain networks under attack: Robustness properties and the impact of lesions. Brain139(12), 3063–3083. https://doi.org/10.1093/brain/aww194 (2016).
Hart, M. G. et al. Global effects of focal brain tumors on functional complexity and network robustness: A prospective cohort study. Neurosurgery84(6), 1201–1213. https://doi.org/10.1093/neuros/nyy378 (2019).
Aerts, H. & Marinazzo, D. ”BTC_preop”. OpenNeuro, 2022. https://doi.org/10.18112/openneuro.ds001226.v5.0.0.
Shen, X. et al. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage82, 403–415. https://doi.org/10.1016/j.neuroimage.2013.05.081 (2013).
Thomas Yeo, B. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol.106(3), 1125–1165. https://doi.org/10.1152/jn.00338.2011 (2011).
Deco, G. et al. Novel Intrinsic Ignition Method Measuring Local-Global Integration Characterizes Wakefulness and Deep Sleep. In: eneuro 4.5 (Sept. 2017), ENEURO.0106–17.2017. ISSN: 2373-2822. https://doi.org/10.1523/ENEURO.0106-17.2017.
Diniz, C.P. et al. CANTAB object recognition and language tests to detect aging cognitive decline: an exploratory comparative study. In: Clinical Interventions in Aging (Dec. 2014), p 37. ISSN: 1178-1998. https://doi.org/10.2147/CIA.S68186.
Gau, S.S.-F. & Huang, W.-L. Rapid visual information processing as a cognitive endophenotype of attention deficit hyperactivity disorder. Psychol. Med.44(2), 435–446. https://doi.org/10.1017/S0033291713000640 (2014).
Grent-‘t-Jong, T. & Woldorff, M. G. Timing and Sequence of Brain Activity in Top-Down Control of Visual-Spatial Attention. In: PLoS Biology 5.1 (Jan 2, 2007). Ed. by Leslie Ungerleider, e12. ISSN: 1545-7885. https://doi.org/10.1371/journal.pbio.0050012.
Raichle, M. E. The Brain’s Default Mode Network. Annu. Rev. Neurosci.38(1), 433–447. https://doi.org/10.1146/annurev-neuro-071013-014030 (2015).
Buckner, R. L. & DiNicola, L. M. The brain’s default network: Updated anatomy, physiology and evolving insights. Nat. Rev. Neurosci.20(10), 593–608. https://doi.org/10.1038/s41583-019-0212-7 (2019).
Pasquini, L. et al. Brain functional connectivity in low- and high-grade gliomas: Differences in network dynamics associated with tumor grade and location. Cancers 14.14 , 3327. https://doi.org/10.3390/cancers14143327.
Whittle, I. R. et al. Meningiomas. Lancet363(9420), 1535–1543. https://doi.org/10.1016/S0140-6736(04)16153-9 (2004).
Derks, J., Reijneveld, J. C. & Douw, L. Neural network alterations underlie cognitive deficits in brain tumor patients. Curr. Opin. Oncol.26(6), 627–633. https://doi.org/10.1097/CCO.0000000000000126 (2014).
Capper, D. Addressing diffuse glioma as a systemic brain disease with single-cell analysis. Arch. Neurol.69(4), 523. https://doi.org/10.1001/archneurol.2011.2910 (2012).
Osswald, M. et al. Brain tumour cells interconnect to a functional and resistant network. Nature528(7580), 93–98. https://doi.org/10.1038/nature16071 (2015).
Buckingham, S. C. et al. Glutamate release by primary brain tumors induces epileptic activity. Nat. Med.17(10), 1269–1274. https://doi.org/10.1038/nm.2453 (2011).
Sontheimer, H. A role for glutamate in growth and invasion of primary brain tumors. J. Neurochem.105(2), 287–295. https://doi.org/10.1111/j.1471-4159.2008.05301.x (2008).
Aerts, H. et al. Modeling Brain Dynamics in Brain Tumor Patients Using the Virtual Brain. In: eneuro 5.3 (May 2018), ENEURO.0083–18.2018. ISSN: 2373-2822. https://doi.org/10.1523/ENEURO.0083-18.2018.
Duffau, H. Functional recovery after surgical resection of low grade gliomas in eloquent brain: hypothesis of brain compensation. J. Neurol. Neurosurg. Psychiatry74(7), 901–907. https://doi.org/10.1136/jnnp.74.7.901 (2003).
Marek, S. et al. Reproducible brain-wide association studies require thousands of individuals. Nature603(7902), 654–660. https://doi.org/10.1038/s41586-022-04492-9 (2022).
Aerts, H. et al. Modeling brain dynamics after tumor resection using the virtual brain. Neuroimage213, 116738. https://doi.org/10.1016/j.neuroimage.2020.116738 (2020).
Aerts, H. et al. Pre- and post-surgery brain tumor multimodal magnetic resonance imaging data optimized for large scale computational modelling. Sci. Data9(1), 676. https://doi.org/10.1038/s41597-022-01806-4 (2022).
Jenkinson, M. et al. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage17(2), 825–841. https://doi.org/10.1006/nimg.2002.1132 (2002).
Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp.17(3), 143–155. https://doi.org/10.1002/hbm.10062 (2002).
Abraham, A. et al. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. https://doi.org/10.3389/fninf.2014.00014 (2014).
Chang, L. NeuroVault Image 395091: Shen et al., 2013 k=268 Whole-Brain Parcellation. https://identifiers.org/neurovault.image:395091. Contributed by Luke Chang on July 6, 2020. Collection: Naturalistic Data Analysis Course Images. Description: Nifti image of the Shen et al., 2013 k=268 whole-brain parcellation. Parcellation was created applying groupwise graph-theory-based parcellation to resting state data. July 2020.
NeuroData. Neuroparc. 2020. https://doi.org/10.17605/OSF.IO/67A3T.
Deco, G. & Kringelbach, M. L. Hierarchy of Information Processing in the Brain: A Novel ‘Intrinsic Ignition Framework. Neuron94(5), 961–968. https://doi.org/10.1016/j.neuron.2017.03.028 (2017).
Escrichs, A. et al. Characterizing the dynamical complexity underlying meditation. Front. Syst. Neurosci.13, 27. https://doi.org/10.3389/fnsys.2019.00027 (2019).
Panda, R. et al. Whole-brain analyses indicate the impairment of posterior integration and thalamo-frontotemporal broadcasting in disorders of consciousness. Hum. Brain Mapp.44(11), 4352–4371. https://doi.org/10.1002/hbm.26386 (2023).
Padilla, N. et al. Disrupted resting-sate brain network dynamics in children born extremely preterm. Cereb. Cortex33(13), 8101–8109. https://doi.org/10.1093/cercor/bhad101 (2023).
Glerean, E. et al. Functional magnetic resonance imaging phase synchronization as a measure of dynamic functional connectivity. Brain Connect.2(2), 91–101. https://doi.org/10.1089/brain.2011.0068 (2012).
Tagliazucchi, E. et al. Criticality in large-scale brain fMRI dynamics unveiled by a novel point process analysis. Front. Physiol. https://doi.org/10.3389/fphys.2012.00015 (2012).
Faul, F. et al. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behavior Research Methods 41(4), 1149–1160. (2009) https://doi.org/10.3758/BRM.41.4.1149.
Lazic, S. E. The problem of pseudoreplication in neuroscientific studies: Is it affecting your analysis?. BMC Neurosci.11(1), 5. https://doi.org/10.1186/1471-2202-11-5 (2010).
Funding
A.J., I.M., and G.P. were supported by Grant PID2021-122136OB-C22 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”, and AGAUR research support grant (ref. 2021 SGR 01035) funded by the Department of Research and Universities of the Generalitat of Catalunya. A.E. was supported by the project eBRAIN-Health - Actionable Multilevel Health Data (id 101058516), funded by the EU Horizon Europe and by the European Union’s Horizon Europe research and innovation programme under the Marie Sklodowska-Curie Actions (ID: 101207460, NEUROCONTRA, HORIZON-MSCA-2024-PF-01-01). G.D. is supported by Grant PID2022-136216NB-I00 funded by MICIU/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”, ERDF, EU, Project NEurological MEchanismS of Injury, and Sleep-like cellular dynamics (NEMESIS) (ref. 101071900) funded by the EU ERC Synergy Horizon Europe, and AGAUR research support grant (ref. 2021 SGR 00917) funded by the Department of Research and Universities of the Generalitat of Catalunya.
Author information
Authors and Affiliations
Contributions
A.J. and G.P. prepared the initial draft of the manuscript. All authors contributed to manuscript revision and corrections. A.J. and A.E. performed the experiments. A.J., G.P., and G.D. conceptualized the study. A.J. and I.M. implemented the methodology.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Juncà, A., Escrichs, A., Martín, I. et al. Impact of meningioma and glioma on whole-brain dynamics. Sci Rep (2026). https://doi.org/10.1038/s41598-026-35140-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-35140-1