Abstract
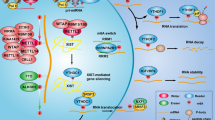
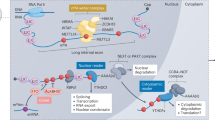
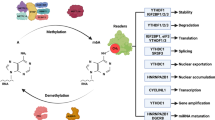
N6-methyladenosine (m6A) RNA methylation plays a crucial role in tumorigenesis. However, the specific role of m6A modifications in the malignant progression of papillary thyroid carcinoma (PTC) without autoimmune thyroid disease (AITD) remains unclear. We analyzed a randomly selected subset of three pairs from a total cohort of 26 pairs of cancerous and para-cancerous tissues from patients with PTC without AITD to investigate global m6A levels and the gene expression of key factors driving m6A methylation. Our results revealed a significant increase in global m6A methylation in cancerous tissues, accompanied by upregulation of the m6A “reader” gene IGF2BP2. Of the 486 upregulated and 39 downregulated genes identified in cancerous tissues, most of the top-enriched pathways were associated with activated genes and contributed to cancer progression. A significant protein–protein interaction between IGF2BP2 and several key cancer-related genes, particularly FN1 and LAMB3, was observed. Moreover, 313 mRNAs, 55 lncRNAs, and 8 ncRNAs exhibited significant m6A methylation differences, overlapping with differentially expressed cancer-associated genes, particularly NUM, which was recently identified as a potential biomarker for PTC. These findings underscore the importance of m6A-related mechanisms and functions in PTC without AITD development and suggest that FN1-, NMU-, and LAMB3-associated pathways may be potential therapeutic targets and molecular mechanisms for this disease.
Similar content being viewed by others
Data availability
The datasets generated during the current study are available in the Gene Expression Omnibus (GEO) repository, under accession number [GSE287426].
References
Pizzato, M. et al. The epidemiological landscape of thyroid cancer worldwide: GLOBOCAN estimates for incidence and mortality rates in 2020. Lancet Diabetes Endocrinol. 10, 264–272. https://doi.org/10.1016/s2213-8587(22)00035-3 (2022).
Pace-Asciak, P., Russell, J. O. & Tufano, R. P. Surgical treatment of thyroid cancer: Established and novel approaches. Best Pract. Res. Clin. Endocrinol. Metab. 37, 101664. https://doi.org/10.1016/j.beem.2022.101664 (2023).
Boucai, L., Zafereo, M. & Cabanillas, M. E. Thyroid cancer: A review. JAMA 331, 425–435. https://doi.org/10.1001/jama.2023.26348 (2024).
Zhang, Y. et al. Targeted therapy and drug resistance in thyroid cancer. Eur. J. Med. Chem. 238, 114500. https://doi.org/10.1016/j.ejmech.2022.114500 (2022).
Morton, L. M. et al. Radiation-related genomic profile of papillary thyroid carcinoma after the Chernobyl accident. Science (New York, N.Y.) https://doi.org/10.1126/science.abg2538 (2021).
Zhang, L. et al. Molecular basis and targeted therapy in thyroid cancer: Progress and opportunities. Biochimica et Biophysica Acta (BBA) Rev. Cancer 1878, 188928. https://doi.org/10.1016/j.bbcan.2023.188928 (2023).
White, M. G. et al. Epigenetic alterations and canonical pathway disruption in papillary thyroid cancer: A genome-wide methylation analysis. Ann. Surg. Oncol. 23, 2302–2309. https://doi.org/10.1245/s10434-016-5185-4 (2016).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. https://doi.org/10.1038/nchembio.687 (2011).
Zhuang, H. et al. The role of m6A methylation in therapy resistance in cancer. Mol. Cancer 22, 91. https://doi.org/10.1186/s12943-023-01782-2 (2023).
Zhou, Z. et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol. Cancer 19, 104. https://doi.org/10.1186/s12943-020-01216-3 (2020).
Sun, H., Li, K., Liu, C. & Yi, C. Regulation and functions of non-m(6)A mRNA modifications. Nat. Rev. Mol. Cell Biol. 24, 714–731. https://doi.org/10.1038/s41580-023-00622-x (2023).
Zheng, L. et al. The emerging roles of the interaction between m6A modification and c-Myc in driving tumorigenesis and development. J. Cell. Physiol. 237, 2758–2769. https://doi.org/10.1002/jcp.30733 (2022).
Liu, L. et al. Insights into N6-methyladenosine and programmed cell death in cancer. Mol. Cancer 21, 32. https://doi.org/10.1186/s12943-022-01508-w (2022).
Li, C., Xie, P., Lv, K., Yang, Q. & Mou, Y. METTL3-mediated m6A modification of hsa_circ_0131922 attenuates the progression of papillary thyroid cancer by regulating the p53 pathway. Endocr. Metab. Immune Disord. Drug Targets. https://doi.org/10.2174/0118715303290063240530053252 (2024).
Zheng, X. et al. N(6)-methyladenosine reader IGF2BP3 as a prognostic Biomarker contribute to malignant progression of glioma. Transl. Cancer Res. 12, 992–1005. https://doi.org/10.21037/tcr-23-449 (2023).
Xu, X. et al. The emerging roles of N6-methyladenosine RNA modifications in thyroid cancer. Eur. J. Med. Res. https://doi.org/10.1186/s40001-023-01382-2 (2023).
Zhou, X. et al. The m6A methyltransferase METTL3 drives thyroid cancer progression and lymph node metastasis by targeting LINC00894. Cancer Cell Int. https://doi.org/10.1186/s12935-024-03240-5 (2024).
Lin, S. et al. METTL3-Induced miR-222-3p upregulation inhibits STK4 and promotes the malignant behaviors of thyroid carcinoma cells. J. Clin. Endocrinol. Metab. 107, 474–490. https://doi.org/10.1210/clinem/dgab480 (2022).
Allegri, L., Baldan, F., Molteni, E., Mio, C. & Damante, G. Role of m6A RNA methylation in thyroid cancer cell lines. Int. J. Mol. Sci. 23, 11516. https://doi.org/10.3390/ijms231911516 (2022).
He, J. et al. METTL3 restrains papillary thyroid cancer progression via m6A/c-Rel/IL-8-mediated neutrophil infiltration. Mol. Ther. 29, 1821–1837. https://doi.org/10.1016/j.ymthe.2021.01.019 (2021).
Zhu, Y. et al. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 13, 358. https://doi.org/10.1038/s41419-022-04817-6 (2022).
Huang, C., Duan, Z., Chen, B., Xia, H. & Wang, G. LncRNA LINC00969 modified by METTL3 attenuates papillary thyroid cancer progression in an m6A-dependent manner. Adv. Clin. Exp. Med. 34, 0–0. https://doi.org/10.17219/acem/188367 (2024).
Ferrari, S. M. et al. Thyroid autoimmune disorders and cancer. Semin. Cancer Biol. 64, 135–146. https://doi.org/10.1016/j.semcancer.2019.05.019 (2020).
Wu, S. et al. N6-Methyladenosine and rheumatoid arthritis: A comprehensive review. Front. Immunol. https://doi.org/10.3389/fimmu.2021.731842 (2021).
Xu, Y., Liu, W. & Ren, L. Emerging roles and mechanism of m6A methylation in rheumatoid arthritis. Biomed. Pharmacother. 170, 116066. https://doi.org/10.1016/j.biopha.2023.116066 (2024).
Lv, X. et al. RNA methylation in systemic lupus erythematosus. Front. Cell Dev. Biol. https://doi.org/10.3389/fcell.2021.696559 (2021).
Li, Z. et al. N6-methyladenosine in inflammatory diseases: Important actors and regulatory targets. Gene 936, 149125. https://doi.org/10.1016/j.gene.2024.149125 (2025).
Li, L. et al. RNA methylation: A potential therapeutic target in autoimmune disease. Int. Rev. Immunol. 43, 160–177. https://doi.org/10.1080/08830185.2023.2280544 (2024).
Tang, L. et al. Emerging perspectives of RNA N6-methyladenosine (m6A) modification on immunity and autoimmune diseases. Front. Immunol. https://doi.org/10.3389/fimmu.2021.630358 (2021).
Mo, K. et al. Targeting hnRNPC suppresses thyroid follicular epithelial cell apoptosis and necroptosis through m6A-modified ATF4 in autoimmune thyroid disease. Pharmacol. Res. 196, 106933. https://doi.org/10.1016/j.phrs.2023.106933 (2023).
Song, R.-h., Du, P., Gao, C.-q., Liu, X.-r. & Zhang, J.-a. METTL3 Is involved in the development of Graves’ disease by inducing SOCS mRNA m6A modification. Front. Endocrinol. https://doi.org/10.3389/fendo.2021.666393 (2021).
Li, X. et al. The essential role of N6-methyladenosine RNA methylation in complex eye diseases. Genes Diseases 10, 505–520. https://doi.org/10.1016/j.gendis.2022.05.008 (2023).
Kanehisa, M., Sato, Y, Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462, https://doi.org/10.1093/nar/gkv1070 (2016).
Kanehisa M & S, G. KEGG kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Xia, J., Shi, Y. & Chen, X. New insights into the mechanisms of the extracellular matrix and its therapeutic potential in anaplastic thyroid carcinoma. Sci. Rep. https://doi.org/10.1038/s41598-024-72020-y (2024).
Qin, Y., Ding, W., Wu, X. & Qin, D. Dabrafenib inhibits Egr-1-mediated adhesion of thyroid cancer cells to pulmonary microvascular endothelium. J. Biochem. Mol. Toxicol. https://doi.org/10.1002/jbt.70060 (2024).
Zhang, N., Ding, C., Zuo, Y., Peng, Y. & Zuo, L. N6-methyladenosine and neurological diseases. Mol. Neurobiol. 59, 1925–1937. https://doi.org/10.1007/s12035-022-02739-0 (2022).
Qin, J. et al. IGF2BP3 drives gallbladder cancer progression by m6A-modified CLDN4 and inducing macrophage immunosuppressive polarization. Transl. Oncol. 37, 101764. https://doi.org/10.1016/j.tranon.2023.101764 (2023).
Zhang, Y. et al. An analysis of differentially expressed and differentially m6A-modified transcripts in soybean roots treated with lead. J. Hazard. Mater. 453, 131370. https://doi.org/10.1016/j.jhazmat.2023.131370 (2023).
Shao, Y., Duan, X., Zhao, X., Lv, Z. & Li, C. Global N6-methyladenosine methylation analysis reveals the positive correlation between m6A modification and mRNA abundance during Apostichopus japonicus disease development. Dev. Comp. Immunol. 133, 104434. https://doi.org/10.1016/j.dci.2022.104434 (2022).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. https://doi.org/10.1038/s41556-018-0045-z (2018).
Zhang, Y. et al. YTHDF3as a prognostic predictive biomarker of thyroid cancer and its correlation with immune infiltration. BMC Cancer 23, 882. https://doi.org/10.1186/s12885-023-11361-9 (2023).
Han, H. et al. Expression and prognostic value of m6A RNA methylation-related genes in thyroid cancer. Iran J. Public Health. 52, 1902–1916 (2023).
Sa, R., Guo, M., Liu, D. & Guan, F. AhR antagonist promotes differentiation of papillary thyroid cancer via regulating circSH2B3/miR-4640-5P/IGF2BP2 axis. Front. Pharmacol. 12, 795386. https://doi.org/10.3389/fphar.2021.795386 (2021).
Sa, R. et al. Targeting IGF2BP2 promotes differentiation of radioiodine refractory papillary thyroid cancer via destabilizing RUNX2 mRNA. Cancers 14, 1268. https://doi.org/10.3390/cancers14051268 (2022).
Wang, W., Ding, Y., Zhao, Y. & Li, X. m6A reader IGF2BP2 promotes lymphatic metastasis by stabilizing DPP4 in papillary thyroid carcinoma. Cancer Gene Ther. 31, 285–299. https://doi.org/10.1038/s41417-023-00702-2 (2023).
Wang, X. et al. Identification and validation of m6A RNA methylation regulators with clinical prognostic value in Papillary thyroid cancer. Cancer Cell Int. 20, 203. https://doi.org/10.1186/s12935-020-01283-y (2020).
Yan, K. et al. Spatial transcriptomics reveals prognosis-associated cellular heterogeneity in the papillary thyroid carcinoma microenvironment. Clin. Transl. Med. 14, e1594. https://doi.org/10.1002/ctm2.1594 (2024).
Chen, Z. et al. Single-cell RNA sequencing revealed a 3-gene panel predicted the diagnosis and prognosis of thyroid papillary carcinoma and associated with tumor immune microenvironment. Front. Oncol. 12, 862313. https://doi.org/10.3389/fonc.2022.862313 (2022).
Wang, Y. et al. Upregulated LAMB3 increases proliferation and metastasis in thyroid cancer. Onco. Targets. Ther. 11, 37–46. https://doi.org/10.2147/ott.s149613 (2017).
Pan, L., Zhang, L., Fu, J., Shen, K. & Zhang, G. Integrated transcriptome sequencing and weighted gene co-expression network analysis reveals key genes of papillary thyroid carcinomas. Heliyon 10, e27928. https://doi.org/10.1016/j.heliyon.2024.e27928 (2024).
Liu, Y. et al. Long noncoding RNA GAS5 targeting miR-221-3p/cyclin-dependent kinase inhibitor 2B axis regulates follicular thyroid carcinoma cell cycle and proliferation. Pathobiology 88, 289–300. https://doi.org/10.1159/000513338 (2021).
Chen, Z. et al. METTL3 promotes cellular senescence of colorectal cancer via modulation of CDKN2B transcription and mRNA stability. Oncogene 43, 976–991. https://doi.org/10.1038/s41388-024-02956-y (2024).
Liu, L. et al. Identification of key genes and pathways of thyroid cancer by integrated bioinformatics analysis. J. Cell. Physiol. 234, 23647–23657. https://doi.org/10.1002/jcp.28932 (2019).
Zeng, Y. M. W. et al. Identification and validation of eight estrogen-related genes for predicting prognosis of papillary thyroid cancer. Aging 15, 1668–1684 (2023).
S., K. TMPRSS4, a type II transmembrane serine protease, as a potential therapeutic target in cancer. Exp. Mol. Med. 55, 716–724. https://doi.org/10.1038/s12276-023-00975-5 (2023).
Xu, X., Sun, T. & Jing, J. TMPRSS4 is a novel biomarker and correlated with immune infiltration in thyroid carcinoma. BMC Endocr. Disord. 22, 280. https://doi.org/10.1186/s12902-022-01203-3 (2022).
Zhao, X.-F., Yang, Y.-S., Gao, D.-Z. & Park, Y. K. TMPRSS4 overexpression promotes the metastasis of colorectal cancer and predicts poor prognosis of stage III–IV colorectal cancer. Int. J. Biol. Mark. 36, 23–32. https://doi.org/10.1177/17246008211046368 (2021).
Oh, B. S. et al. Inhibition of TMPRSS4 mediated epithelial-mesenchymal transition is critically involved in antimetastatic effect of melatonin in colorectal cancers. Phytother. Res. 35, 4538–4546. https://doi.org/10.1002/ptr.7156 (2021).
Chen, Y. et al. LncRNA TGFB2-OT1 promotes progression and angiogenesis in hepatocellular carcinoma by dephosphorylating β-catenin. J. Hepatocell. Carcinoma 10, 429–446. https://doi.org/10.2147/jhc.s404008 (2023).
Song, P. et al. A three-lncRNA expression signature associated with the prognosis of gastric cancer patients. Cancer Med. 6, 1154–1164. https://doi.org/10.1002/cam4.1047 (2017).
Acknowledgements
We are grateful to the patients for their help and willingness to participate in this study. This study was supported by the Natural Science Foundation of the Fujian Province (Grant Number: 2022J01222) and the Fujian Provincial Health Commission Youth Scientific Research Project (Grant Number: 2023QNA037).
Funding
This study was funded by Natural Science Foundation of Fujian Province (2022J01222), Fujian Provincial Health Commission Youth Scientific Research Project (2023QNA037).The funding body had no further role in the study design, decision to publish or preparation of manuscript.
Author information
Authors and Affiliations
Contributions
RY designed the experiments. SL, YL, and HZ performed the experiments. ZJ and RY analyzed the data. ZJ and RY prepared an initial draft of the manuscript. HZ and RY revised the manuscript. All the authors commented on and approved the final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, Z., Luo, S., Lin, Y. et al. Correlations of m6A methylation-related mRNAs with thyroid cancer. Sci Rep (2026). https://doi.org/10.1038/s41598-026-35712-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-35712-1