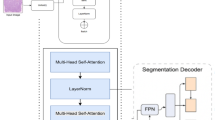
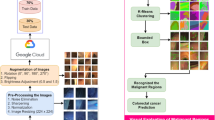
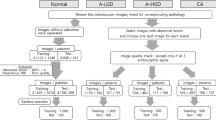
Abstract
Accurate gland segmentation in colorectal cancer histopathology is crucial, but the scarcity of pixel-level annotations limits robust model development. This study aims to develop a highly accurate gland segmentation method that leverages weakly labeled data, specifically image-level labels, to overcome the need for extensive pixel-level annotations. We propose a novel three-stage framework that uniquely combines self-supervised fine-tuning of the DINOv2 vision transformer, attention-based pseudo-label generation, and a boundary-aware loss function. Initially, an off-the-shelf DINOv2 encoder is fine-tuned on a large unlabeled dataset of histopathology images. This fine-tuned encoder is then integrated into a classification network equipped with an attention mechanism, which is trained using image-level labels to generate initial pseudo-labels via attention maps. These maps are refined through blending, thresholding, and Conditional Random Field (CRF) post-processing. Finally, a segmentation network, employing the same fine-tuned encoder and a lightweight decoder, is trained using these refined pseudo-labels and a boundary-aware loss. Ablation studies demonstrated the significant benefit of the fine-tuned encoder and the comprehensive post-processing steps for pseudo-label generation. Further studies confirmed the effectiveness of the boundary-aware loss in improving segmentation accuracy. Our method achieved superior performance on the GlaS dataset compared to several state-of-the-art methods, including both fully supervised and weakly supervised approaches, demonstrating higher F1-score, Object Dice, and lower Object Hausdorff distance. This approach effectively addresses the challenge of limited pixel-level annotations by utilizing more readily available image-level data, offering a promising solution for improved colorectal cancer diagnosis. The proposed framework shows potential for generalization to other histopathology image analysis tasks.
Similar content being viewed by others
Data availability
The GlaS dataset and IMP-CRS-2024 dataset are publicly available from the MICCAI 2015 Gland Segmentation Challenge (https://warwick.ac.uk/fac/cross_fac/tia/data/glascontest/) and IMP Diagnostics and INESC TEC (https://open-datasets.inesctec.pt/NQ3sxFMZ/), respectively.
Code availability
To ensure reproducibility and to support future research, the implementation code has been made publicly available on Zenodo at https://zenodo.org/records/17677517.
References
Bray, F. et al. Global cancer statistics 2022: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 74, 229–263, https://doi.org/10.3322/caac.21834 (2024).
Bokhorst, J.-M. et al. Deep learning for multi-class semantic segmentation enables colorectal cancer detection and classification in digital pathology images. Scientific Reports 13, 8398. https://doi.org/10.1038/s41598-023-35491-z (2023).
Song, A. H. et al. Artificial intelligence for digital and computational pathology. Nature Reviews Bioengineering 1, 930–949. https://doi.org/10.1038/s44222-023-00096-8 (2021).
Bilal, M., Nimir, M., Snead, D., Taylor, G. S. & Rajpoot, N. Role of ai and digital pathology for colorectal immuno-oncology. British journal of cancer 128, 3–11. https://doi.org/10.1038/s41416-022-01986-1 (2023).
Davri, A. et al. Deep learning on histopathological images for colorectal cancer diagnosis: A systematic review. Diagnostics 12, https://doi.org/10.3390/diagnostics12040837 (2022).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015, 234–241 (2015).
Chen, H., Qi, X., Yu, L. & Heng, P.-A. Dcan: Deep contour-aware networks for accurate gland segmentation. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2487–2496, https://doi.org/10.1109/CVPR.2016.273 (2016).
Xu, Y. et al. Gland instance segmentation using deep multichannel neural networks. IEEE Transactions on Biomedical Engineering 64, 2901–2912. https://doi.org/10.1109/TBME.2017.2686418 (2017).
Graham, S. et al. Mild-net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Medical Image Analysis 52, 199–211. https://doi.org/10.1016/j.media.2018.12.001 (2019).
Zhang, S. et al. Venet: Variational energy network for gland segmentation of pathological images and early gastric cancer diagnosis of whole slide images. Computer Methods and Programs in Biomedicine 250, 108178. https://doi.org/10.1016/j.cmpb.2024.108178 (2024).
Sun, M., Wang, J., Gong, Q. & Huang, W. Enhancing gland segmentation in colon histology images using an instance-aware diffusion model. Computers in Biology and Medicine 166, 107527. https://doi.org/10.1016/j.compbiomed.2023.107527 (2023).
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A. & Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2921–2929 (2016).
Ahn, J. & Kwak, S. Learning pixel-level semantic affinity with image-level supervision for weakly supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 4981–4990 (2018).
Ahn, J., Cho, S. & Kwak, S. Weakly supervised learning of instance segmentation with inter-pixel relations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2209–2218 (2019).
Jo, S. & Yu, I.-J. Puzzle-cam: Improved localization via matching partial and full features. arXiv preprint arXiv:2101.11253 (2021).
Lee, J., Kim, E. & Yoon, S. Anti-adversarially manipulated attributions for weakly and semi-supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 4071–4080 (2021).
Ru, L., Zhan, Y., Yu, B. & Du, B. Learning affinity from attention: End-to-end weakly-supervised semantic segmentation with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2022).
Zhang, B. et al. Contain: A contrastive approach for object-level weakly supervised semantic segmentation. arXiv preprint (2023).
Xu, G. et al. Camel: A weakly supervised learning framework for histopathology image segmentation. 2019 IEEE/CVF International Conference on Computer Vision (ICCV) 10681–10690, https://doi.org/10.1109/ICCV.2019.01078 (2019).
Chang, Y.-T. et al. Weakly-supervised semantic segmentation via sub-category exploration. CVPR 8991–9000, https://doi.org/10.48550/arXiv.2008.01183 (2020).
Li, Y., Yu, Y., Zou, Y., Xiang, T. & Li, X. Online easy example mining for weakly-supervised gland segmentation from histology images. Medical Image Computing and Computer Assisted Intervention - MICCAI 2022, 578–587 (2022).
Song, J., Yun, S., Yoon, S., Kim, J. & Lee, S. Ep-sam: Weakly supervised histopathology segmentation via enhanced prompt with segment anything. ArXiv arXiv:abs/2410.13621 (2024).
He, K. et al. Masked autoencoders are scalable vision learners (2021). arXiv:2111.06377.
Caron, M. et al. Emerging properties in self-supervised vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) (2021).
Zhou, J. et al. ibot: Image BERT pre-training with online tokenizer. arXiv preprint arXiv:2111.07832 (2021).
Chen, R. J. et al. Towards a general-purpose foundation model for computational pathology. Nature Medicine (2024).
Zimmermann, E. et al. Virchow: A foundation model for pathology. arXiv preprint (2024).
Xu, H. et al. Whole-slide foundation model for digital pathology. Nature Medicine (2024).
Filiot, A. et al. Scaling self-supervised learning for histopathology with masked image modeling. arXiv preprint (2024).
Bioptimus. H-optimus-0: An open-source foundation model for pathology. arXiv preprint arXiv:2411.08950 (2024).
Gui, J. et al. Dinov2: Learning robust visual features without supervision. arXiv preprint (2023).
Ilse, M., Tomczak, J. M. & Welling, M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning (ICML), vol. 80, 2127–2136 (PMLR, 2018).
Krähenbühl, P. & Koltun, V. Efficient inference in fully connected CRFs with Gaussian edge potentials. In Advances in Neural Information Processing Systems (NeurIPS) (2011).
Kervadec, H. et al. Boundary loss for highly unbalanced segmentation. In Medical Imaging with Deep Learning (MIDL) (2019).
Oliveira, S. P. et al. Cad systems for colorectal cancer from wsi are still not ready for clinical acceptance. Scientific Reports 11, 1–15. https://doi.org/10.1038/s41598-021-93746-z (2021).
Neto, P. C. et al. imil4path: A semi-supervised interpretable approach for colorectal whole-slide images. Cancers 14, 2489 (2022).
Neto, P. C. et al. An interpretable machine learning system for colorectal cancer diagnosis from pathology slides. npj Precision Oncology 8, 56, https://doi.org/10.1038/s41698-024-00539-4 (2024).
Sirinukunwattana, K. et al. Gland segmentation in colon histology images: The glas challenge contest. Medical Image Analysis 35, 489–502. https://doi.org/10.1016/j.media.2016.08.008 (2017).
Macenko, M. et al. A method for normalizing histology slides for quantitative analysis. 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 1107–1110, https://doi.org/10.1109/ISBI.2009.5193250 (2009).
Paszke, A., Gross, S., Massa, F., Lerer, A. & Chintala, S. Pytorch: An imperative style, high-performance deep learning library. ArXiv https://doi.org/10.48550/arXiv.1912.01703 (2019).
Pinckaers, H. & Litjens, G. J. S. Neural ordinary differential equations for semantic segmentation of individual colon glands. ArXiv arXiv:abs/1910.10470 (2019).
Acknowledgements
This work was supported by Natural Science Foundation of Shanghai under grant number 25ZR1401273.
Author information
Authors and Affiliations
Contributions
H.W. and Y.W. conceived the experiment(s), H.W. conducted the experiment(s), D.H. and C.L. analysed the results. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical Statement
This study utilized publicly available and de-identified datasets (GlaS, IMP-CRS-2024, CRAG). Ethical review and approval were not required for the use of these pre-existing datasets.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wen, H., Wu, Y., Huang, D. et al. Weakly supervised colorectal gland segmentation through self-supervised learning and attention-based pseudo-labeling. Sci Rep (2026). https://doi.org/10.1038/s41598-026-36256-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-36256-0