Abstract
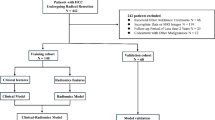
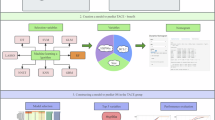
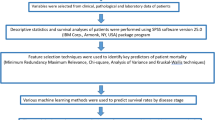
This study aims to construct a robust artificial intelligence (AI) model to predict early recurrence of hepatocellular carcinoma (HCC) following surgical resection, leveraging clinical blood biomarkers, pathological parameters, and MRI-derived features. We included 240 hepatectomy patients from two medical centers, collecting clinical blood biomarkers, MRI features, and postoperative pathological data. Feature reduction was conducted using Spearman correlation and the least absolute shrinkage and selection operator (LASSO) regression. Predictive models were constructed using five machine learning algorithms and validated on an external dataset. The models were subsequently compared. The ExtraTrees, XGBoost, and LightGBM models exhibited high predictive performance in the training set, with AUCs of 0.816 (95% CI 0.748–0.884), 0.978 (95% CI 0.958–0.998), and 0.898 (95% CI 0.846–0.950), respectively. In the validation set, their AUC values were 0.759 (95% CI 0.641–0.876), 0.789 (95% CI 0.684–0.894), and 0.760 (95% CI 0.650–0.869). Decision curve analysis indicated favorable net benefits for predicting early recurrence across all three models. Tumor margin and age were identified as significant factors, showing strong associations with early recurrence. This study developed AI model utilizing clinical blood biomarkers, MRI features, and pathological information to predict early recurrence of HCC after surgery. The models demonstrated good predictive performance and showed clinical applicability in predicting early recurrence, potentially assisting clinicians in identifying high-risk patients, guiding individualized surveillance, and optimizing postoperative management. However, inherent biases in this retrospective study necessitate further research for validation and refinement.
Similar content being viewed by others
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- AI:
-
Artificial intelligence
- HCC:
-
Hepatocellular carcinoma
- LASSO:
-
The least absolute shrinkage and selection operator
- AFP:
-
Alpha-fetoprotein levels
- NEUT:
-
Neutrophi
- LY:
-
Lymphocyte
- NLR:
-
Neutrophil-to-lymphocyte ratio
- PLT:
-
Platelet count
- ALB:
-
Albumin
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
- PA:
-
Prealbumin
- GGT:
-
Gamma-glutamyl transferase
- TBIL:
-
Total bilirubin
- DBIL:
-
Direct bilirubin
- PT:
-
Prothrombin time
- INR:
-
International normalized ratio
- HBP:
-
Hepatobiliary phase
- BCLC:
-
Barcelona clinical liver cancer
- MVI:
-
Microvascular invasion
- ES:
-
Edmondson–Steiner
- AUC:
-
Area under the receiver operating characteristic curve
- DCA:
-
Decision curve analysis
References
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 7 (1), 6. https://doi.org/10.1038/s41572-020-00240-3 (2021).
Zhang, Z. H. et al. Role of microvascular invasion in early recurrence of hepatocellular carcinoma after liver resection: A literature review. Asian J. Surg. 47 (5), 2138–2143. https://doi.org/10.1016/j.asjsur.2024.02.115 (2024).
Shi, S., Zhao, Y. X., Fan, J. L., Chang, L. Y. & Yu, D. X. Development and external validation of a nomogram including body composition parameters for predicting early recurrence of hepatocellular carcinoma after hepatectomy. Acad. Radiol. 30 (12), 2940–2953. https://doi.org/10.1016/j.acra.2023.05.022 (2023).
Cheng, Z. et al. Risk factors and management for early and late intrahepatic recurrence of solitary hepatocellular carcinoma after curative resection. HPB 17 (5), 422–427. https://doi.org/10.1111/hpb.12367 (2015).
Nevola, R. et al. Predictors of early and late hepatocellular carcinoma recurrence. World J. Gastroenterol. 29 (8), 1243–1260. https://doi.org/10.3748/wjg.v29.i8.1243 (2023).
Zeng, J. et al. Development of a machine learning model to predict early recurrence for hepatocellular carcinoma after curative resection. Hepatobiliary Surg. Nutr. 11 (2), 176–187. https://doi.org/10.21037/hbsn-20-466 (2022).
Gao, W. et al. A predictive model integrating deep and radiomics features based on gadobenate dimeglumine-enhanced MRI for postoperative early recurrence of hepatocellular carcinoma. Radiol. Med. 127 (3), 259–271. https://doi.org/10.1007/s11547-021-01445-6 (2022).
Zhang, Z. et al. Hepatocellular carcinoma: radiomics nomogram on Gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging. 19 (1), 22–32. https://doi.org/10.1186/s40644-019-0209-5 (2019).
Ji, G. W. et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: a multi-institutional study. EBioMedicine 50, 156–165. https://doi.org/10.1016/j.ebiom.2019.10.057 (2019).
Yan, M. et al. Deep learning nomogram based on gd-EOB-DTPA MRI for predicting early recurrence in hepatocellular carcinoma after hepatectomy. Eur. Radiol. 33 (7), 4949–4961. https://doi.org/10.1007/s00330-023-09419-0 (2023).
Miranda, J. et al. Current status and future perspectives of radiomics in hepatocellular carcinoma. World J. Gastroenterol. 29 (1), 43–60. https://doi.org/10.3748/wjg.v29.i1.43 (2023).
Zheng, C. et al. Gut Microbiome as a biomarker for predicting early recurrence of < span style=font-variant:Small-caps;>HBV -related hepatocellular carcinoma. Cancer Sci. 114 (12), 4717–4731. https://doi.org/10.1111/cas.15983 (2023).
Zhou, S. L. et al. Genomic sequencing identifies WNK2 as a driver in hepatocellular carcinoma and a risk factor for early recurrence. J. Hepatol. 71 (6), 1152–1163. https://doi.org/10.1016/j.jhep.2019.07.014 (2019).
Fu, Y. et al. Combining a machine-learning derived 4-lncRNA signature with AFP and TNM stages in predicting early recurrence of hepatocellular carcinoma. BMC Genom. 24 (1), 89–104. https://doi.org/10.1186/s12864-023-09194-8 (2023).
Dong, B. et al. Development of a machine learning-based model to predict prognosis of alpha-fetoprotein-positive hepatocellular carcinoma. J. Transl. Med. 22 (1), 455–465. https://doi.org/10.1186/s12967-024-05203-w (2024).
Wang, X. et al. Clinical-radiological characteristic for predicting ultra-early recurrence after liver resection in solitary hepatocellular carcinoma patients. J. Hepatocell Carcinoma. 10, 2323–2335. https://doi.org/10.2147/JHC.S434955 (2023).
European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 69 (1), 182–236. https://doi.org/10.1016/j.jhep.2018.03.019 (2018).
Kang, I. et al. Subclassification of microscopic vascular invasion in hepatocellular carcinoma. Ann. Surg. 274 (6), e1170–e1178. https://doi.org/10.1097/SLA.0000000000003781 (2021).
Yan, Y. et al. Radiomic analysis based on Gd-EOB-DTPA enhanced MRI for the preoperative prediction of Ki-67 expression in hepatocellular carcinoma. Acad. Radiol. 31 (3), 859–869. https://doi.org/10.1016/j.acra.2023.07.019 (2024).
Shah, S. A. et al. Factors associated with early recurrence after resection for hepatocellular carcinoma and outcomes. J. Am. Coll. Surg. 202 (2), 275–283. https://doi.org/10.1016/j.jamcollsurg.2005.10.005 (2006).
Zeng, J. et al. Development of a machine learning model to predict early recurrence for hepatocellular carcinoma after curative resection. HepatoBiliary Surg. Nutr. 11 (2). https://doi.org/10.21037/hbsn-20-466 (2022).
Wei, Y., Pei, W., Qin, Y., Su, D. & Liao, H. Preoperative MR imaging for predicting early recurrence of solitary hepatocellular carcinoma without microvascular invasion. Eur. J. Radiol. 138, 109663. https://doi.org/10.1016/j.ejrad.2021.109663 (2021).
Kawashima, J. et al. Development and validation of a preoperative risk prediction model for severe complications and very early recurrence after liver resection for hepatocellular carcinoma. Surgery 185, 109527. https://doi.org/10.1016/j.surg.2025.109527 (2025).
Ariizumi, S. et al. A non-smooth tumor margin in the hepatobiliary phase of Gadoxetic acid disodium (Gd‐EOB‐DTPA) ‐enhanced magnetic resonance imaging predicts microscopic portal vein invasion, intrahepatic metastasis, and early recurrence after hepatectomy in patients with hepatocellular carcinoma. J. Hepato Biliary Pancreat. 18 (4), 575–585. https://doi.org/10.1007/s00534-010-0369-y (2011).
Zhang, L. et al. The role of preoperative dynamic contrast-enhanced 3.0-T MR imaging in predicting early recurrence in patients with early-Stage hepatocellular carcinomas after curative resection. Front. Oncol. 9, 1336. https://doi.org/10.3389/fonc.2019.01336 (2019).
Chou, C. T. et al. Prediction of microvascular invasion of hepatocellular carcinoma: preoperative CT and histopathologic correlation. Am. J. Roentgenol. 203 (3), W253–W259. https://doi.org/10.2214/AJR.13.10595 (2014).
Chartampilas, E. et al. Current imaging diagnosis of hepatocellular carcinoma. Cancers 14 (16), 3997. https://doi.org/10.3390/cancers14163997 (2022).
Liu, C. et al. A K-nearest neighbor model to predict early recurrence of hepatocellular carcinoma after resection. J. Clin. Transl. Hepatol. 10 (4), 600–607. https://doi.org/10.14218/JCTH.2021.00348 (2022).
Wang, H. et al. A novel nomogram predicting the early recurrence of hepatocellular carcinoma patients after R0 resection. Front. Oncol. 13, 1133807. https://doi.org/10.3389/fonc.2023.1133807 (2023).
Yen, Y. H. et al. A simple model to predict early recurrence of hepatocellular carcinoma after liver resection. Langenbecks Arch. Surg. 409 (1), 261. https://doi.org/10.1007/s00423-024-03449-y (2024).
Giuffrè, M. et al. Predictors of hepatocellular carcinoma early recurrence in patients treated with surgical resection or ablation treatment: a single-center experience. Diagnostics 12 (10), 2517. https://doi.org/10.3390/diagnostics12102517 (2022).
Ryu, J. M. et al. Prognostic impact of elevation of cancer antigen 15 – 3 (CA15-3) in patients with early breast cancer with normal serum CA15-3 level. J. Breast Cancer. 26 (2), 126–136. https://doi.org/10.4048/jbc.2023.26.e17 (2023).
Zheng, C. et al. Nomogram based on clinical and preoperative CT features for predicting the early recurrence of combined hepatocellular-cholangiocarcinoma: a multicenter study. Radiol. med. 128 (12), 1460–1471. https://doi.org/10.1007/s11547-023-01726-2 (2023).
Wang, J. et al. 3D MR elastography of hepatocellular carcinomas as a potential biomarker for predicting tumor recurrence. Magn. Reson. Imaging. 49 (3), 719–730. https://doi.org/10.1002/jmri.26250 (2019).
Zang, Y., Long, P., Wang, M., Huang, S. & Chen, C. Development and validation of prognostic nomograms in patients with hepatocellular carcinoma: a population-based study. Future Oncol. 17 (36), 5053–5066. https://doi.org/10.2217/fon-2020-1065 (2021).
Qin, C., Gao, Y., Li, J., Huang, C. & He, S. Predictive effects of preoperative serum CA125 and AFP levels on post-hepatectomy survival in patients with hepatitis B-related hepatocellular carcinoma. Oncol. Lett. 21 (6), 487–500. https://doi.org/10.3892/ol.2021.12748 (2021).
Zhang, Z. Y., Guan, J., Wang, X. P., Hao, D. S. & Zhou, Z. Q. Outcomes of adolescent and young patients with hepatocellular carcinoma after curative liver resection: a retrospective study. World J. Surg. Oncol. 20 (1), 210–217. https://doi.org/10.1186/s12957-022-02658-3 (2022).
Zeng, J. et al. Prognosis factors of young patients undergoing curative resection for hepatitis B virus-related hepatocellular carcinoma: a multicenter study. Cancer Manag. Res. 12, 6597–6606. https://doi.org/10.2147/CMAR.S261368 (2020).
Chen, J. P. et al. Pre-operative enhanced magnetic resonance imaging combined with clinical features predict early recurrence of hepatocellular carcinoma after radical resection. World J. Gastrointest. Oncol. 16 (4), 1192–1203. https://doi.org/10.4251/wjgo.v16.i4.1192 (2024).
Romeo, M. et al. Clinical applications of artificial intelligence (AI) in human cancer: is it time to update the diagnostic and predictive models in managing hepatocellular carcinoma (HCC)? Diagnostics 15 (3), 252–274. https://doi.org/10.3390/diagnostics15030252 (2025).
Seven, İ. et al. Predicting hepatocellular carcinoma survival with artificial intelligence. Sci. Rep. 15 (1), 6226. https://doi.org/10.1038/s41598-025-90884-6 (2025).
Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B. 58 (1), 267–288. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x (1996).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 (1), 1–22 (2010).
Feng, L. et al. Predicting overall survival in hepatocellular carcinoma patients via a combined MRI radiomics and pathomics signature. Transl. Oncol. 51, 102174. https://doi.org/10.1016/j.tranon.2024.102174 (2025).
Jiang, W. et al. Pathomics signature for prognosis and chemotherapy benefits in stage III colon cancer. JAMA Surg. 159 (5), 519–529. https://doi.org/10.1001/jamasurg.2023.8015 (2024).
Chen, D. et al. Prognostic and predictive value of a pathomics signature in gastric cancer. Nat. Commun. 13 (1), 6903. https://doi.org/10.1038/s41467-022-34703-w (2022).
Chen, T., Guestrin, C. & XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 785–794. https://doi.org/10.1145/2939672.2939785 (2016).
Funding
This study was supported by the National Natural Science Foundation of China, Grant/Award Number: 82060310.
Author information
Authors and Affiliations
Contributions
L.F.: Conceptualization, formal analysis, visualization, writing original draft, investigation, software. N.L.: Conceptualization, data curation, formal analysis, investigation, validation, visualization, software, writing original draft. F.R.: Investigation, data curation, validation. X.Z.: Data curation, validation. X.P.: Data curation, validation. X.L.: Data curation, validation. L.F.: Data curation, validation. L.L.: Methodology, project administration, writing review and editing, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, L., Luo, N., Ruan, F. et al. Machine learning integrating MRI and clinical features predicts early recurrence of hepatocellular carcinoma after resection. Sci Rep (2026). https://doi.org/10.1038/s41598-026-36261-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-36261-3