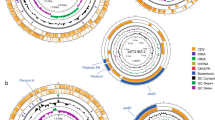
Abstract
This study developed new real-time PCR assays for six lactic acid bacteria (LAB): Ligilactobacillus agilis, Limosilactobacillus fermentum, Lactobacillus johnsonii, Ligilactobacillus salivarius, Pediococcus pentosaceus, and Weissella cibaria for their application in the food industry. For each target bacterium, a novel primer/probe set was designed in a conserved and species-specific genomic region, with proper length (13 ~ 30nt), Tm (probes: 68 ~ 70℃; primers: 58 ~ 60℃), GC content (30 ~ 80%), and amplicon length (50 ~ 150 bp). The Gibbs free energy (ΔG, an indicator for stability of secondary structures) of potential hairpins, self-dimers, and cross-dimers of the primers and probes adhered largely to the recommended values with minor deviations. BLAST analysis verified the conservation and specificity of each target sequence. After establishing the reaction mixture and the thermal procedure, each new assay was preliminarily evaluated in inclusivity, specificity, amplification efficiency, and precision. All tested samples of each target bacterium generated positive results. All tested non-target bacterial samples yielded negative results. Amplification efficiency of each assay, measured using 10-fold serial dilutions of a positive sample, was 95 ~ 100%. Precision (repeatability and reproducibility) of each assay generated relative standard deviations (RSDs) < 2%. Collectively, these newly developed assays predictably have high inclusivity, specificity, amplification efficiency, and precision, which need more comprehensive validation before industrial application.
Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
References
Shehata, H. R., Hassane, B. & Newmaster, S. G. Real-time PCR methods for identification and stability monitoring of Bifidobacterium longum subsp. longum UABl-14 during shelf life. Front. Microbiol. 15, 1360241. https://doi.org/10.3389/fmicb.2024.1360241 (2024).
Khattab, R. A., Ahmed, N. A., Ragab, Y. M. & Rasmy, S. A. Bacteria producing antimicrobials against Clostridium difficile isolated from human stool. Anaerobe 63, 102206. https://doi.org/10.1016/j.anaerobe.2020.102206 (2020).
Kumar, S. et al. Evaluation of the techno-functional properties of lactobacilli strains originated from Bos indicus and Bubalus bubalis calves for probiotic potential. Int. Microbiol. 28, 1649–1668. https://doi.org/10.1007/s10123-025-00641-y (2025).
Feng, M. et al. Lactobacillus agilis SNF7 presents excellent antibacteria and anti-inflammation properties in mouse diarrhea induced by Escherichia coli. Int. J. Mol. Sci. 25, 13660. https://doi.org/10.3390/ijms252413660 (2024).
Yang, Y. et al. Understanding Ligilactobacillus salivarius from probiotic properties to omics technology: A review. Foods 13, 895. https://doi.org/10.3390/foods13060895 (2024).
de Freire, L. Limosilactobacillus fermentum strains as novel probiotic candidates to promote host health benefits and development of biotherapeutics: A comprehensive review. Probiotics Antimicrob. Proteins. 16, 1483–1498. https://doi.org/10.1007/s12602-024-10235-1 (2024).
Zhou, J. et al. Unveiling the potential of Lactobacillus Johnsonii in digestive diseases: a comprehensive review. Front. Microbiol. 16 https://doi.org/10.3389/fmicb.2025.1508382 (2025).
Jiang, S., Cai, L., Lv, L. & Li, L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell. Fact. 20 https://doi.org/10.1186/s12934-021-01537-y (2021).
Qi, Y. et al. Pediococcus pentosaceus: screening and application as probiotics in food processing. Front. Microbiol. 12, 762467. https://doi.org/10.3389/fmicb.2021.762467 (2021).
Kim, E., Yang, S. M., Kim, I. S. & Kim, H. Y. Identification of novel molecular targets for Weissella species-specific real-time PCR based on pangenome analysis. Appl. Microbiol. Biotechnol. 106, 4157–4168. https://doi.org/10.1007/s00253-022-12003-z (2022).
Kim, H. B. et al. Development of real-time PCR assay to specifically detect 22 Bifidobacterium species and subspecies using comparative genomics. Front. Microbiol. 11, 2087. https://doi.org/10.3389/fmicb.2020.02087 (2020).
Kim, E., Lee, G. Y., Yang, S. M. & Kim, H. Y. Rapid and accurate on-site identification of Lactobacillus delbrueckii subspecies in dairy products using direct polymerase chain reaction with microfluidic chip. Lwt 179, 114635. https://doi.org/10.1016/j.lwt.2023.114635 (2023).
Zamanpour, S., Eraghi, V., Hashemi, M., Ram, M. & Afshari, A. Optimizing real-time PCR for accurate identification and quantification of common probiotic bacteria in Iranian probiotic dairy products. Indian J. Microbiol. 65, 1891–1905. https://doi.org/10.1007/s12088-024-01325-3 (2024).
Kim, E., Yang, S. M., Kim, D. & Kim, H. Y. Real-time PCR method for qualitative and quantitative detection of Lactobacillus sakei group species targeting novel markers based on bioinformatics analysis. Int. J. Food Microbiol. 355, 109335. https://doi.org/10.1016/j.ijfoodmicro.2021.109335 (2021).
Kim, E., Kim, D. S., Yang, S. M. & Kim, H. Y. The accurate identification and quantification of six Enterococcus species using quantitative polymerase chain reaction based novel DNA markers. Lwt 166, 113769. https://doi.org/10.1016/j.lwt.2022.113769 (2022).
Gaspar, C. et al. Development and validation of a new one step multiplex-PCR assay for the detection of ten Lactobacillus species. Anaerobe 59, 192–200. https://doi.org/10.1016/j.anaerobe.2019.06.004 (2019).
Kim, E. et al. Design of PCR assays to specifically detect and identify 37 Lactobacillus species in a single 96 well plate. BMC Microbiol. 20, 96. https://doi.org/10.1186/s12866-020-01781-z (2020).
Applied Biosystems. Thermofisher Primer Express Version 3.0 Getting Started Guide. https://documents.thermofisher.com/TFS-Assets/LSG/manuals/cms_041902.pdf (Accessed 28 Sept 2025).
Benchling, P. Design. https://test.benchling.com/primer-design-for-pcr (Accessed 28 Sept 2025).
Premier Biosoft. PCR Primer Design Guidelines. http://www.premierbiosoft.com/tech_notes/PCR_Primer_Design.html (Accessed 28 Sept 2025).
Kralik, P. & Ricchi, M. A. Basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 8, 108. https://doi.org/10.3389/fmicb.2017.00108 (2017).
Xiang, X. et al. Rapid identification of novel specific molecular targets for PCR detection of four Enterococcus species. Lwt 173, 114356. https://doi.org/10.1016/j.lwt.2022.114356 (2023).
Shehata, H. R. et al. Guidelines for validation of qualitative real-time PCR methods for molecular diagnostic identification of probiotics. J. AOAC Int. 102, 1774–1778. https://doi.org/10.5740/jaoacint.18-0320 (2019).
Svec, D., Tichopad, A., Novosadova, V., Pfaffl, M. W. & Kubista, M. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantification. 3, 9–16. https://doi.org/10.1016/j.bdq.2015.01.005 (2015).
Dung, T. T. N. et al. Development and validation of multiplex real-time PCR for simultaneous detection of six bacterial pathogens causing lower respiratory tract infections and antimicrobial resistance genes. BMC Infect. Dis. 24, 164. https://doi.org/10.1186/s12879-024-09028-2 (2024).
Downs, S. L., Madhi, S. A., van der Merwe, L., Nunes, M. C. & Olwagen, C. P. Optimization of a high-throughput nanofluidic real-time PCR to detect and quantify of 15 bacterial species and 92 Streptococcus pneumoniae serotypes. Sci. Rep. 13, 4588. https://doi.org/10.1038/s41598-023-31820-4 (2023).
Broeders, S. et al. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 37, 115–126. https://doi.org/10.1016/j.tifs.2014.03.008 (2014).
Funding
This research was supported by Key Scientific Research Platforms and Projects for Universities in Guangdong Province (Grant Number: 2024GCZX011), Youth Science Fund Project of National Natural Science Foundation of China (Grant Number: 32502380), Guangdong Provincial Key Discipline Construction Program for Enhancing Research Capacity (Grant Number: 2024ZDJS089), Foshan Self-Financed Science and Technology Innovation Project (Grant Number: 2420001004679), Guangzhou College of Technology and Business Scientific Research Project (Grant Number: KYYB202431), Practical Quality Engineering Construction Project of Guangzhou College of Technology and Business (Grant Number: SYKC2024042).
Author information
Authors and Affiliations
Contributions
Conceptualization, S.-J. Li, C. Yang, G. Zhang and S. Ou; methodology, S.-J. Li and G. Zhang; software, S.-J. Li; validation, B. Cui, W. Li, F. Lu and L. Yuan; formal analysis, L. Han, Y. Xie, X. Sun, Y. Zheng and L. Qian; investigation, K. Luo, S. He, W. Lu, K. Xu, X. Zhu; resources, S.-J. Li, Y. Zheng, and G. Zhang.; data curation, S.-J. Li; writing—original draft preparation, S.-J. Li; writing—review and editing, L. Han, Y. Xie, X. Sun, Y. Zheng and L. Qian; visualization, S.-J. Li; supervision, S.-J. Li; project administration, L. Han, Y. Xie, Y. Zheng, C. Yang and S. Ou; funding acquisition, L. Han, Y. Xie, Y. Zheng, C. Yang and S. Ou.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, SJ., Cui, B., Li, W. et al. Development and preliminary evaluation of real-time PCR assays for six lactic acid bacteria. Sci Rep (2026). https://doi.org/10.1038/s41598-026-37047-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-37047-3