Abstract
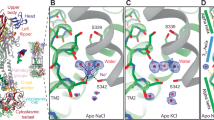
Tyrosylprotein sulfotransferases (TPSTs) catalyze O-sulfation of tyrosine residues on secreted and membrane proteins, but the molecular basis for their stimulation by metal ions remains unclear. We determined the structures of the catalytic domain of human TPST2 with PAP and Na+ (1.75 Å) or Mn2+ (2.00 Å) bound and identified two conserved octahedral metal-binding sites. Anomalous diffraction at metal absorption edges confirmed the identity of the bound metals and demonstrated specific Mn2+ binding. The Na+- and Mn2+-bound structures closely superimposed, suggesting activation without large conformational changes. Structural comparison with the apo structure and ensemble refinement revealed differences in local dynamics around the metal binding sites. The flexible α3-helix and α12-α13 loop in the apo structure were stabilized by Na+ binding and further rigidified by Mn2+ binding. These findings support an activation-by-ordering mechanism in which Na+ binding generates a pre-activated state, with Mn2+ subsequently establishing a catalytically competent ordering that lowers the entropic barrier at the active-site entrance. This framework reconciles longstanding biochemical observations and suggests that Mn2+ availability within the Golgi can tune TPST2-dependent signaling.
Similar content being viewed by others
Data availability
The coordinates and structural factors for TPST2 Na and TPST2 Mn have been deposited in the Protein Data Bank under accession codes 9WWE and 9WWF (PDB DOI: https://doi.org/10.2210/pdb9WWE/pdb and https://doi.org/10.2210/pdb9WWF/pdb). The source data underlying the graphs presented in the paper are provided in Supplementary Data 1. All other data are available from the corresponding author upon reasonable request.
6. References
Lee, R. & Huttner, W. Tyrosine-O-sulfated proteins of PC12 pheochromocytoma cells and their sulfation by a tyrosylprotein sulfotransferase. J. Biol. Chem. 258, 11326–11334 (1983).
Baeuerle, P. A. & Huttner, W. B. Tyrosine sulfation is a trans-Golgi-specific protein modification. J. Cell. Biol. 105, 2655–2664 (1987).
Huttner, W. B. Tyrosine sulfation and the secretory pathway. Annu. Rev. Physiol. 50, 363–376 (1988).
Lipmann, F. Biological sulfate activation and transfer: studies on a mechanism of group activation and its role in biosynthesis are described. Science 128, 575–580 (1958).
Yang, Y. S. et al. Tyrosine sulfation as a protein post-translational modification. Molecules 20, 2138–2164 (2015).
Hille, A., Braulke, T., von FIGURA, K. & Huttner, W. B. Occurrence of tyrosine sulfate in proteins–a balance sheet: 1. Secretory and lysosomal proteins. Eur. J. Biochem. 188, 577–586 (1990).
Hille, A. & Huttner, W. B. Occurrence of tyrosine sulfate in proteins–a balance sheet: 2. Membrane proteins. Eur. J. Biochem. 188, 587–596 (1990).
Kehoe, J. W. & Bertozzi, C. R. Tyrosine sulfation: a modulator of extracellular protein–protein interactions. Chem. Biol. 7, R57–R61 (2000).
Huttner, W. B. Sulphation of tyrosine residues—a widespread modification of proteins. Nature 299, 273–276 (1982).
Moore, K. L. The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 278, 24243–24246 (2003).
Stewart, V. & Ronald, P. C. Sulfotyrosine residues: interaction specificity determinants for extracellular protein–protein interactions. J. Biol. Chem. 298, 102232 (2022).
Ouyang, Y., Lane, W. S. & Moore, K. L. Tyrosylprotein sulfotransferase: purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins. Proc. Natl. Acad. Sci. USA. 95, 2896–2901 (1998).
Beisswanger, R. et al. Existence of distinct tyrosylprotein sulfotransferase genes: molecular characterization of tyrosylprotein sulfotransferase-2. Proc. Natl. Acad. Sci. USA. 95, 11134–11139 (1998).
Ouyang, Y. B. & Moore, K. L. Molecular cloning and expression of human and mouse tyrosylprotein sulfotransferase-2 and a tyrosylprotein sulfotransferase homologue in caenorhabditis elegans. J. Biol. Chem. 273, 24770–24774 (1998).
Kakuta, Y., Pedersen, L. G., Pedersen, L. C. & Negishi, M. Conserved structural motifs in the sulfotransferase family. Trends Biochem. Sci. 23, 129–130 (1998).
Teramoto, T. et al. Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction. Nat. Commun. 4, 1572 (2013).
Tanaka, S. et al. Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1. Sci. Rep. 7, 8776 (2017).
Lee, R. & Huttner, W. B. (Glu62, Ala30, Tyr8)n serves as high-affinity substrate for tyrosylprotein sulfotransferase: a Golgi enzyme. Proc. Natl. Acad. Sci. USA 82, 6143–6147 (1985).
Niehrs, C. & Huttner, W. B. Purification and characterization of tyrosylprotein sulfotransferase. EMBO J. 9, 35–42 (1990).
Bundgaard, J. R., Vuust, J. & Rehfeld, J. F. New consensus features for tyrosine O-sulfation determined by mutational analysis. J. Biol. Chem. 272, 21700–21705 (1997).
Yoshimura, M. et al. Crystal structure of tick tyrosylprotein sulfotransferase reveals the activation mechanism of the tick anticoagulant protein Madanin. J Biol. Chem. 300 (2024).
Westmuckett, A. D., Hoffhines, A. J., Borghei, A. & Moore, K. L. Early postnatal pulmonary failure and primary hypothyroidism in mice with combined TPST-1 and TPST-2 deficiency. Gen. Comp. Endocrinol. 156, 145–153 (2008).
Borghei, A. et al. Targeted disruption of tyrosylprotein sulfotransferase-2, an enzyme that catalyzes post-translational protein tyrosine O-sulfation, causes male infertility. J. Biol. Chem. 281, 9423–9431 (2006).
Sasaki, N. et al. A mutation in Tpst2 encoding tyrosylprotein sulfotransferase causes dwarfism associated with hypothyroidism. Mol. Endocrinol. 21, 1713–1721 (2007).
Mishiro, E., Sakakibara, Y., Liu, M. C. & Suiko, M. Differential enzymatic characteristics and tissue-specific expression of human TPST-1 and TPST-2. J. Biochem. 140, 731–737 (2006).
Oh, Y. et al. Genome-wide CRISPR screening identifies tyrosylprotein sulfotransferase-2 as a target for augmenting anti-PD1 efficacy. Mol. Cancer. 23, 155 (2024).
Cai, X. et al. Inhibition of the SLC35B2-TPST2 axis of tyrosine sulfation attenuates the growth and metastasis of pancreatic ductal adenocarcinom. Cell. Mol. Gastroenterol. Hepatol. 16, 473–495 (2023).
Ujiie, K. et al. Specific recognition mechanism of an antibody to sulfated tyrosine and its potential use in biological research. J. Biol. Chem. 301 (2025).
Kasinathan, C., Rizwan, M., Slomiany, A. & Slomiany, B. L. Effect of sofalcone on tyrosylprotein sulfotransferase. Gen. Pharmacol. 25, 1017–1020 (1994).
Byrne, D. P. et al. New tools for evaluating protein tyrosine sulfation: tyrosylprotein sulfotransferases (TPSTs) are novel targets for RAF protein kinase inhibitors. Biochem. J. 475, 2435–2455 (2018).
Gucwa, M. et al. CMM—An enhanced platform for interactive validation of metal binding sites. Protein Sci. 32, e4525 (2023).
Putignano, V., Rosato, A., Banci, L. & Andreini, C. MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 46, D459–D464 (2018).
Lewis, S. et al. Scalable emulation of protein equilibrium ensembles with generative deep learning. Science 389, eadv9817 (2025).
Das, S. et al. Manganese mapping using a fluorescent Mn2+ sensor and Nanosynchrotron X-ray fluorescence reveals the role of the golgi apparatus as a manganese storage site. Inorg. Chem. 58 (20), 13724–13732 (2019).
Potelle, S. et al. Glycosylation abnormalities in Gdt1p/TMEM165 deficient cells result from a defect in golgi manganese homeostasis. Hum. Mol. Genet. 25 (8), 1489–1500 (2016).
Potelle, S. et al. Manganese-induced turnover of TMEM165. Biochem. J. 474 (9), 1481–1493 (2017).
Jankauskas, S. S. et al. Insights into molecular and cellular functions of the golgi calcium/manganese-proton antiporter TMEM165. J. Biol. Chem. 300 (8), 107567 (2024).
Kabsch, W. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125–132 (2010).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. Sect. D Struct. Biol. 75, 861–877 (2019).
Agirre, J. et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr. Sect. D Struct. Biol. 79, 449–461 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Schrodinger, L. The PyMOL molecular graphics system. Version 1, 8 (2015).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Troshin, P. V., Procter, J. B. & Barton, G. J. Java bioinformatics analysis web services for multiple sequence alignment—JABAWS. MSA Bioinformatics. 27, 2001–2002 (2011).
Troshin, P. V. et al. JABAWS 2.2 distributed web services for bioinformatics: protein disorder, conservation and RNA secondary structure. Bioinformatics 34, 1939–1940 (2018).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Burnley, B. T., Afonine, P. V., Adams, P. D. & Gros, P. Modelling dynamics in protein crystal structures by ensemble refinement. Elife 1, e00311 (2012).
Song, X. et al. Accurate prediction of protein structural flexibility by deep learning integrating intricate atomic structures and Cryo-EM density information. Nat. Commun. 15, 5538 (2024).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods. 19, 679–682 (2022).
Jin, M., Yang, J., Park, J., Kim, H. & Eom, S. H. Structure of MICU from non-metazoan dictyostelium discoideum reveals unique characteristics. Commun. Biol. 8, 782 (2025).
Park, S. B. et al. Discovery of (4-Phenyl-cyclohexyl) acetate-Derived tyrosylprotein sulfotransferase 2 (TPST2) inhibitors with potent anti-tumor activity for immuno-oncology applications. J. Med. Chem. (2025).
Acknowledgements
We gratefully acknowledge the staff at beamlines BL-5 C and 11 C at the PAL for their kind help with data collection. This research was supported by the National Research Foundation (NRF) of the Korean government (NRF-2021R1A2C1006267) and by the Bio&Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (RS-2024-00344154 and RS-2024-00440614).
Funding
This research was supported by the National Research Foundation (NRF) of the Korean government (NRF-2021R1A2C1006267) and by the Bio&Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (RS-2024-00344154 and RS-2024-00440614).
Author information
Authors and Affiliations
Contributions
S.H.E. and M.J. conceived the study and organized experiments; M.J. performed most experiments; C.N., J.Y., and H.K. contributed to X-ray diffraction experiments and data analysis; S.B.P. contributed to enzyme activity assays and analysis; S.H.E. and Y.C.K. provided advice and guidance. All authors contributed to the interpretation of the results and preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jin, M., Noh, C., Yang, J. et al. Structural characterization of metal binding in human tyrosylprotein sulfotransferase 2, TPST2. Sci Rep (2026). https://doi.org/10.1038/s41598-026-37189-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-37189-4