Abstract
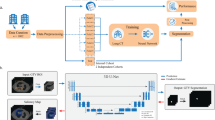
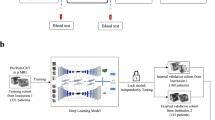
Rare diseases are often difficult to diagnose, and their scarcity also makes it challenging to develop deep learning models for them due to limited large-scale datasets. Anterior mediastinal tumors—including thymoma and thymic carcinoma—represent such rare entities. A few diagnostic support systems for these tumors have been proposed; however, no prior studies have tested them across multiple institutions, and clinically applicable and generalizable models remain lacking. A total of 711 computed tomography (CT) images were collected from 136 hospitals, each from a different patient with pathologically proven anterior mediastinal tumors (339 males, 372 females). Of these, 485 images were used for training, 62 for tuning, and 164 for external testing. The external testing dataset comprised CT images from 121 unique institutions not involved in the other datasets. A 3D U-Net-based model was trained on the training dataset, and the model with the best performance on the tuning dataset was selected. This model was then evaluated on the external testing dataset for its segmentation and detection performance across different institutions. Based on the reference standards provided by board-certified diagnostic radiologists, the trained model achieved average Dice scores of 0.82, Intersection over Union (IoU) of 0.72, Precision of 0.85, and Recall of 0.82 for tumor segmentation at the CT-image level. The free-response receiver operating characteristic curve—derived from lesion-wise IoU thresholds—demonstrated high sensitivity and a low false-positive rate for tumor detection. Even under a stricter IoU threshold of 0.50, the model maintained a sensitivity of 0.87 with only 0.61 false positives per scan. Our model achieved clinically applicable segmentation and detection performance for anterior mediastinal tumors, demonstrating broad generalizability across 121 institutions and overcoming the data-scarcity challenges inherent to such rare diseases.
Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to patient privacy concerns but are available from the corresponding author on reasonable request. The trained model weights are available at https://huggingface.co/hirwatan/FFSCS.
References
Tomiyama, N. et al. Anterior mediastinal tumors: Diagnostic accuracy of CT and MRI. Eur. J. Radiol. 69, 280–288. https://doi.org/10.1016/j.ejrad.2007.10.002 (2009).
Takahashi, K. & Al-Janabi, N. J. Computed tomography and magnetic resonance imaging of mediastinal tumors. J. Magn. Reson. Imaging 32, 1325–1339. https://doi.org/10.1002/jmri.22377 (2010).
Marom, E. M. Advances in thymoma imaging. J. Thorac. Imaging 28, 69–80. https://doi.org/10.1097/RTI.0b013e31828609a0 (2013).
Marx, A. et al. The 2021 WHO Classification of Tumors of the Thymus and Mediastinum: What is new in thymic epithelial, germ cell, and mesenchymal tumors?. J. Thorac. Oncol. 17, 200–213. https://doi.org/10.1016/j.jtho.2021.10.010 (2022).
Tang, R. et al. Pan-mediastinal neoplasm diagnosis via nationwide federated learning: A multicentre cohort study. Lancet Digit. Health 5, 560–570. https://doi.org/10.1016/S2589-7500(23)00106-1 (2023).
Huang, S. et al. Anterior mediastinal lesion segmentation based on two-stage 3D ResUNet with attention gates and lung segmentation. Front. Oncol. 10, 618357. https://doi.org/10.3389/FONC.2020.618357/BIBTEX (2021).
Zhou, Z. et al. Privacy enhancing and generalizable deep learning with synthetic data for mediastinal neoplasm diagnosis. NPJ Digit. Med. 7, 1–15 (2024).
Huang, S., Yu, H., Ai, D., Ma, G. & Yang, J. CLIP and image integrative prompt for anterior mediastinal lesion segmentation in CT image. 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 99, 3315–3318. https://doi.org/10.1109/BIBM62325.2024.10822165 (2024).
Yamada, S. et al. Automatic assessment of disproportionately enlarged subarachnoid-space hydrocephalus from 3D MRI using two deep learning models. Front. Aging Neurosci. 16, 1362637. https://doi.org/10.3389/fnagi.2024.1362637 (2024).
Tejani, A. S. Checklist for artificial intelligence in medical imaging (CLAIM): 2024 update. Radiol. Artif. Intell. https://doi.org/10.1148/RYAI.240300 (2024).
Carter, B. W. et al. ITMIG classification of mediastinal compartments and multidisciplinary approach to mediastinal masses. Radiographics 37, 413–436. https://doi.org/10.1148/RG.2017160095/ASSET/IMAGES/LARGE/RG.2017160095.FIG20B.JPEG (2017).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015 (eds Navab, N. et al.) 234–241 (Springer, Cham, 2015).
Milletari, F., Navab, N., & Ahmadi, S.-A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE, Stanford, CA, USA https://doi.org/10.1109/3DV.2016.79 (2016).
Aerts, H. J. W. L. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006. https://doi.org/10.1038/ncomms5006 (2014).
Dasegowda, G. et al. No code machine learning: Validating the approach on use-case for classifying clavicle fractures. Clin. Imaging 112, 110207. https://doi.org/10.1016/j.clinimag.2024.110207 (2024).
Walston, S. L. et al. Data set terminology of deep learning in medicine: A historical review and recommendation. Jpn. J. Radiol. 42, 1100–1109. https://doi.org/10.1007/s11604-024-01608-1 (2024).
Santomartino, S. M. et al. Performance and usability of code-free deep learning for chest radiograph classification, object detection, and segmentation. Radiol. Artif. Intell. 5, 220062. https://doi.org/10.1148/ryai.220062 (2023).
Jarrahi, M. H., Memariani, A. & Guha, S. The principles of data-centric AI. Commun. ACM 66(8), 84–92. https://doi.org/10.1145/3571724 (2023).
Yang, Y. et al. Development and validation of contrast-enhanced CT-based deep transfer learning and combined clinical-radiomics model to discriminate thymomas and thymic cysts: A multicenter study. Acad. Radiol. 31, 1615–1628. https://doi.org/10.1016/J.ACRA.2023.10.018 (2024).
Nakajo, M. et al. The efficacy of18F-FDG-PET-based radiomic and deep-learning features using a machine-learning approach to predict the pathological risk subtypes of thymic epithelial tumors. Brit. J. Radiol. https://doi.org/10.1259/BJR.20211050/SUPPL_FILE/BJR.20211050.SUPPL-01.DOCX (2022).
Han, S. et al. Fully automatic quantitative measurement of 18F-FDG PET/CT in thymic epithelial tumors using a convolutional neural network. Clin. Nucl. Med. 47, 590–598. https://doi.org/10.1097/RLU.0000000000004146 (2022).
Yamada, D. et al. Multimodal modeling with low-dose CT and clinical information for diagnostic artificial intelligence on mediastinal tumors: A preliminary study. BMJ Open Respir. Res. 11, 002249. https://doi.org/10.1136/BMJRESP-2023-002249 (2024).
Wang, F. et al. A deep learning model combining circulating tumor cells and radiological features in the multi-classification of mediastinal lesions in comparison with thoracic surgeons: A large-scale retrospective study. BMC Med. 23, 1–11. https://doi.org/10.1186/S12916-025-04104-Z (2025).
Acknowledgements
This work was supported by the National Cancer Center Research and Development Fund (grant number: 2023-A-19). We would like to express our sincere gratitude to Hironori Matsumasa, Keigo Nakamura, and Hokuto Yonezawa from Medical System Research & Development Center, FUJIFILM Corporation, Tokyo, Japan, for the invaluable assistance in data analysis for this study.
Funding
This work was supported by the National Cancer Center Research and Development Fund (grant number: 2023-A-19).
Author information
Authors and Affiliations
Contributions
All authors contributed to the conceptualization and/or design of this study. C.T. performed data curation, formal analysis, investigation, software operation, validation, visualization, and drafted the original manuscript. M.M. performed conceptualization, data curation, formal analysis, investigation, methodology, project administration, software operation, supervision, validation, visualization, and manuscript writing (review and editing). K.K. contributed to conceptualization, investigation, software operation, supervision, validation, visualization, and writing (review and editing). H.M. contributed to investigation, software operation, validation, visualization, and writing (review and editing). R.S. contributed to investigation, validation, and writing (review and editing). A.U. performed data curation, formal analysis, investigation, software operation, supervision, validation, visualization, and manuscript writing (review and editing). Y.G., Y.Y., S.W., M.K., and R.H. participated in manuscript writing (review and editing). M.S. contributed to visualization, investigation, and manuscript writing (review and editing). H.W. was responsible for data curation, conceptualization, funding acquisition, investigation, methodology, project administration, resources, supervision, visualization, and manuscript writing (review and editing). All authors had approved the final version. They take responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
K.K. has received research funding from FUJIFILM Corporation. A.U. has received honoraria for lectures on CT imaging from Canon Medical Systems, Japan; GE Healthcare Pharma, Japan; and Fuji Pharma, Japan. Furthermore, A.U. serves as the Vice President of the Japanese Society of CT Technology and as a Delegate for the Japanese Society of Radiological Technology. Y.G. has received grants or contracts paid to his institution from AIQIVA Services Japan, MSD, Astellas Pharma, AstraZeneca, AbbVie, Amgen, Syneos Health, Sysmex Corporation, CMIC, Novartis Pharma, Bayer Pharmaceuticals, Bristol-Myers Squibb, MedPace Japan, Janssen Pharma, Clinical Research Support Center Kyushu, SATOMI, Ono Pharmaceutical, Daiichi Sankyo, Takeda Pharmaceutical, Chugai Pharmaceutical, NPO Thoracic Oncology Research Group, Eli Lilly Japan, and Preferred Network. He has also received grants or contracts paid to himself from AstraZeneca, AbbVie, Eli Lilly, Pfizer, Bristol Myers Squibb, Ono, Novartis, Kyorin, and Daiichi Sankyo. In addition, Y.G. has received payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing, or educational events from Eli Lilly, Chugai, Taiho, Boehringer Ingelheim, Ono, Bristol Myers Squibb, Pfizer, MSD, Novartis, Merck, and Thermo Fisher. He has served on the monitoring or advisory boards for AstraZeneca, Chugai, Boehringer Ingelheim, Eli Lilly, GlaxoSmithKline, Taiho, Pfizer, Novartis, Kyorin, Guardant Health Inc., Illumina, Daiichi-Sankyo, Merck, MSD, Ono, and Janssen. He also holds a leadership or fiduciary role in Cancer Net Japan and JAMT. M.K. has received grants or contracts from Canon Medical Systems Corporation. M.K. has also received consulting fees from Daiichi-Sankyo Co., Ltd. R.H. has received research funds under contract from Fujifilm Corporation since the initial planning of this work. All other authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Takemura, C., Miyake, M., Kobayashi, K. et al. A clinically applicable and generalizable deep learning model for anterior mediastinal tumors in CT images across multiple institutions. Sci Rep (2026). https://doi.org/10.1038/s41598-026-37504-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-37504-z