Abstract
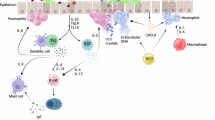
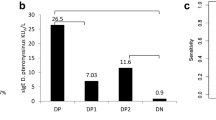
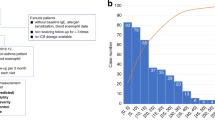
Asthma heterogeneity remains a major barrier in precision medicine. Although elevated total serum immunoglobulin E (IgE) is a hallmark of asthma, even in nonatopic patients, its causal role in asthma pathogenesis is debated. We hypothesized that genetic predisposition to increased IgE defines a distinct asthma endotype. A genome-wide association study of total serum IgE in 1,287 non-asthmatic Japanese adults was used to construct IgE polygenic risk scores (IgE_PRS). Applying IgE_PRS to 745 patients with asthma, we performed cluster analysis incorporating age at onset, total IgE levels, IgE_PRS, and percent predicted forced expiratory volume in 1 s (pFEV1), identifying four distinct adult asthma phenotypes. Notably, one cluster had the highest IgE_PRS and adult-onset-predominant type 2 inflammation. Conversely, the second cluster displayed the highest IgE levels but average IgE_PRS. The remaining two clusters comprised patients with lower IgE_PRS. One cluster was characterized by eosinophilia and smoking-related airflow limitation, whereas the other exhibited a type 2 low phenotype. In a 10-year retrospective cohort, over 30% of newly diagnosed asthma cases fell into the genetically predisposed high-IgE_PRS cluster. These findings reveal a distinct adult-onset-predominant asthma phenotype driven by genetically determined IgE production, offering new avenues for endotype-driven diagnosis and personalized therapy.
Similar content being viewed by others
Data availability
The GWAS genotype data of the non-asthmatic controls and asthma patients collected by our group at the University of Tsukuba cannot be deposited in a public repository, since no consent was obtained for deposition ***. However, these data are available upon request (contact: nhizawa@md.tsukuba.ac.jp) for use in research on inflammatory lung diseases.***.
References
Fuhlbrigge, A. L. & Sharma, S. Unraveling the heterogeneity of asthma: decoding subtypes of asthma. J. Allergy Clin. Immunol. 156, 41–50 (2025).
Burrows, B., Martinez, F. D., Halonen, M., Barbee, R. A. & Cline, M. G. Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl. J. Med. 320, 271–277 (1989).
Habib, N., Pasha, M. A. & Tang, D. D. Current Understanding of asthma pathogenesis and biomarkers. Cells 11 (2022).
Eggel, A., Pennington, L. F. & Jardetzky, T. S. Therapeutic monoclonal antibodies in allergy: targeting IgE, cytokine, and alarmin pathways. Immunol. Rev. 328, 387–411 (2024).
Kashiwakura, J., Otani, I. M. & Kawakami, T. Monomeric IgE and mast cell development, survival and function. Adv. Exp. Med. Biol. 716, 29–46 (2011).
Asai, K. et al. Regulation of mast cell survival by IgE. Immunity 14, 791–800 (2001).
Chen, M., Su, Q. & Shi, Y. Molecular mechanism of IgE-mediated FcεRI activation. Nature 637, 453–460 (2025).
Hanson, B. et al. Atopic disease and Immunoglobulin E in twins reared apart and together. Am. J. Hum. Genet. 48, 873–879 (1991).
Hopp, R. J., Bewtra, A. K., Watt, G. D., Nair, N. M. & Townley, R. G. Genetic analysis of allergic disease in twins. J. Allergy Clin. Immunol. 73, 265–270 (1984).
Jacobsen, H. P., Herskind, A. M., Nielsen, B. W. & Husby, S. IgE in unselected like-sexed monozygotic and dizygotic twins at birth and at 6 to 9 years of age: high but dissimilar genetic influence on IgE levels. J. Allergy Clin. Immunol. 107, 659–663 (2001).
Granada, M. et al. A genome-wide association study of plasma total IgE concentrations in the Framingham heart study. J. Allergy Clin. Immunol. 129, 840–845e821 (2012).
Yatagai, Y. et al. Genome-wide association study for levels of total serum IgE identifies HLA-C in a Japanese population. PLoS One. 8, e80941 (2013).
Kitazawa, H. et al. ORMDL3/GSDMB genotype as a risk factor for early-onset adult asthma is linked to total serum IgE levels but not to allergic sensitization. Allergol. Int. 70, 55–60 (2021).
Moffatt, M. F. et al. A large-scale, consortium-based genomewide association study of asthma. N Engl. J. Med. 363, 1211–1221 (2010).
Sutoh, Y. et al. Genetic predisposition for Immunoglobulin E production explains atopic risk in children: Tohoku medical megabank cohort study. Am. J. Hum. Genet. 112, 1852–1863 (2025).
Lu, H. F. et al. The genome-wide association study of serum IgE levels demonstrated a shared genetic background in allergic diseases. Clin. Immunol. 260, 109897 (2024).
Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017).
Marsh, D. G. et al. Genetics of human immune response to allergens. J. Allergy Clin. Immunol. 65, 322–332 (1980).
Caillat-Zucman, S. How NKG2D ligands trigger autoimmunity? Hum. Immunol. 67, 204–207 (2006).
Thomas, S. A. & Lajoie, S. Complement’s involvement in allergic Th2 immunity: a cross-barrier perspective. J Clin. Invest 135 (2025).
Yatagai, Y. et al. Variants near the HLA complex group 22 gene (HCG22) confer increased susceptibility to late-onset asthma in Japanese populations. J. Allergy Clin. Immunol. 138, 281–283e213 (2016).
Xie, W. et al. Genome-Wide analyses reveal gene influence on HIV disease progression and HIV-1 C acquisition in Southern Africa. AIDS Res. Hum. Retroviruses. 33, 597–609 (2017).
Nalls, M. A. et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 7, e1002113 (2011).
Kato, N., Nakanishi, M. & Hirashima, N. Flotillin-1 regulates IgE receptor-mediated signaling in rat basophilic leukemia (RBL-2H3) cells. J. Immunol. 177, 147–154 (2006).
Dang, C. et al. TCF19 drives a broad transcriptional program that potentiates optimal innate and adaptive functions of antiviral NK cells. Nat. Immunol. 26, 1467–1475 (2025).
Oji, V. et al. Loss of Corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am. J. Hum. Genet. 87, 274–281 (2010).
Li, X. et al. Discoidin domain receptor 1(DDR1) promote intestinal barrier disruption in ulcerative colitis through tight junction proteins degradation and epithelium apoptosis. Pharmacol. Res. 183, 106368 (2022).
Paller, A. S., Spergel, J. M., Mina-Osorio, P. & Irvine, A. D. The atopic March and atopic multimorbidity: many trajectories, many pathways. J. Allergy Clin. Immunol. 143, 46–55 (2019).
Calışkan, M. et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl. J. Med. 368, 1398–1407 (2013).
Lefaudeux, D. et al. U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J. Allergy Clin. Immunol. 139, 1797–1807 (2017).
Konno, S. et al. Distinct phenotypes of smokers with fixed airflow limitation identified by cluster analysis of severe asthma. Ann. Am. Thorac. Soc. 15, 33–41 (2018).
Parnes, J. R. et al. Targeting TSLP in asthma. J. Asthma Allergy. 15, 749–765 (2022).
Tam, V. et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20, 467–484 (2019).
Han, M. et al. Comparison of three multiple allergen simultaneous tests: RIDA allergy screen, MAST optigen, and polycheck allergy. Biomed. Res. Int. 340513 2013 (2013).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal Pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Yatagai, Y. et al. Expression quantitative trait loci for ETV4 and MEOX1 are associated with adult asthma in Japanese populations. Sci. Rep. 11, 18791 (2021).
Kaneko, Y. et al. Asthma phenotypes in Japanese Adults - Their associations with the CCL5 ADRB2 genotypes. Allergol. Int. 62, 113–121 (2013).
Yatagai, Y. et al. Genomewide association study identifies HAS2 as a novel susceptibility gene for adult asthma in a Japanese population. Clin. Exp. Allergy. 44, 1327–1334 (2014).
Ren, J. et al. Asian screening array and Next-Generation sequencing based panels applied to preimplantation genetic testing for Monogenic disorders preclinical workup in 294 families: A retrospective analysis. Prenat Diagn. 44, 1344–1353 (2024).
Choi, S. W., Mak, T. S. & O’Reilly, P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15, 2759–2772 (2020).
Turner, S. et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet Chap. 1, Unit1.19 (2011).
Wolthers, O. D. & Staberg, M. The usefulness of the multiple allergen simultaneous test-chemiluminescent as compared to the Phadia Immunocap IgE test panel system in children and adolescents. Recent. Pat. Inflamm. Allergy Drug Discov. 7, 96–99 (2013).
Toyota, H. et al. Comprehensive analysis of allergen-specific IgE in COPD: mite-specific IgE specifically related to the diagnosis of asthma-COPD overlap. Allergy Asthma Clin. Immunol. 17, 13 (2021).
Kanazawa, J. et al. How important is allergic sensitization as a cause of atopic asthma? Allergol. Int. 67, 292–294 (2018).
Heeringa, J. J. et al. IgE-expressing memory B cells and plasmablasts are increased in blood of children with asthma, food allergy, and atopic dermatitis. Allergy 73, 1331–1336 (2018).
Sakaue, S. et al. Trans-biobank analysis with 676,000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat. Med. 26, 542–548 (2020).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience 8 (2019).
Privé, F., Vilhjálmsson, B. J., Aschard, H. & Blum, M. G. B. Making the most of clumping and thresholding for polygenic scores. Am. J. Hum. Genet. 105, 1213–1221 (2019).
Higgins, J. P. T. et al. VA. Cochrane Handbook for Systematic Reviews of Interventions. version 6.5 ednCochrane, (2024).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Shigemasa, R. et al. Genetic impact of CDHR3 on the adult onset of asthma and COPD. Clin. Exp. Allergy. 50, 1223–1229 (2020).
Tibshirani, R., Hastie, T., Narasimhan, B. & Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U S A. 99, 6567–6572 (2002).
Acknowledgements
We thank Ms. Takako Nakamura for her assistance with the genotyping.
Funding
The authors received no funding for this work.
Author information
Authors and Affiliations
Contributions
T.M., H.M., and N.H. organized and designed the study. T.M., H.M., Y.O., R.S., H.K., Y.Y., and N.H. participated in sample collection and contributed to data analysis.T.M., H.M., and N.H. drafted the manuscript. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved as part of the research project titled “Genetic Predisposition to Inflammatory Obstructive Pulmonary Diseases” (a human genome and gene analysis research project) by the Institutional Review Boards (IRBs) of the University of Tsukuba (IRB No. 136-4) and the University of Tsukuba Hospital (IRB No. H29-294).
Informed consent
Written informed consent was obtained from all participants, and all procedures were conducted in accordance with the Ethical Guidelines for Human Genome/Gene Analysis Research established by the three ministries of the Japanese government.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Matsuda, T., Masuko, H., Ozawa, Y. et al. Genetic predisposition to elevated total immunoglobulin E levels defines a distinct adult-onset-predominant asthma phenotype. Sci Rep (2026). https://doi.org/10.1038/s41598-026-37679-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-37679-5