Abstract
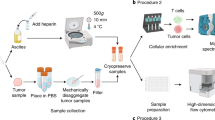
Reprogramming of cellular metabolism is a hallmark of cancer, particularly ovarian cancer (OC), that contributes to rapid cancer growth and survival. However, studies using clinical specimens are limited. To identify metabolic alterations specific to OC, we performed metabolomic analysis of OC and benign ovarian tumors. The relationship between metabolomics and transcriptomics was investigated by transcriptome analysis. Fifty-one patients with OC and three with benign ovarian tumors, diagnosed between 2011 and 2014 using available frozen tissue and plasma specimens, were enrolled at the National Cancer Center Hospital. To identify metabolic alterations, plasma samples from 51 patients with OC and three with benign tumors, along with both cancerous and non-cancerous tissue samples from 44 of the 51 patients with OC, were analyzed using gas chromatography-mass spectrometry. In addition, we performed transcriptomic analysis of cancerous tissues obtained from 39 of the 44 patients with OC. It was not possible to classify patients based on plasma metabolite levels; therefore, the 44 patients with OC were classified into two groups based on metabolite levels: high and low, based on tissue analysis. The group with high metabolite levels had more advanced-stage tumors (P = 0.02). Transcriptome pathway analysis revealed suppression of pathways related to natural killer (NK) cells and immune responses in the group with high metabolite levels. NK cell percentages were lower in the group with high metabolite levels than in the group with low metabolite levels (P = 0.04). Thus, the group with high metabolite levels was associated with advanced stages and a reduced fraction of NK cells, suggesting that high metabolite levels may play a direct or indirect role in immune activity or in the malignant progression of OC.
Similar content being viewed by others
Data availability
Raw RNA sequencing and whole-exome sequencing data have been deposited in the NBDC Human Database under project accession number hum0524 (https://humandbs.dbcls.jp/en/hum0524-v1).
References
Allemani, C. et al. Global surveillance of cancer survival 1995–2009: Analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 385, 977–1010 (2015).
Cusimano, M. C. et al. Ovarian cancer incidence and death in average-risk women undergoing bilateral salpingo-oophorectomy at benign hysterectomy. Am. J. Obstet. Gynecol. 226(220), e221-220 e226 (2022).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J Clin. 70, 7–30 (2020).
Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022).
Zhang, Y., Wang, Y., Zhao, G., Orsulic, S. & Matei, D. Metabolic dependencies and targets in ovarian cancer. Pharmacol. Ther. 245, 108413 (2023).
Tan, Y. et al. Metabolic reprogramming from glycolysis to fatty acid uptake and beta-oxidation in platinum-resistant cancer cells. Nat. Commun. 13, 4554 (2022).
Li, J. et al. Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell 20, 303-314 e305 (2017).
De Martino, M., Rathmell, J. C., Galluzzi, L. & Vanpouille-Box, C. Cancer cell metabolism and antitumour immunity. Nat Rev Immunol. 24, 654–669 (2024).
Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science. 368, eaaw5473 (2020).
Schwartz, L., Seyfried, T., Alfarouk, K. O., Da Veiga Moreira, J. & Fais, S. Out of Warburg effect: An effective cancer treatment targeting the tumor specific metabolism and dysregulated pH. Semin. Cancer Biol. 43, 134–138 (2017).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011).
Jin, Z., Gu, J., Xin, X., Li, Y. & Wang, H. Expression of hexokinase 2 in epithelial ovarian tumors and its clinical significance in serous ovarian cancer. Eur. J. Gynaecol. Oncol. 35, 519–524 (2014).
Chao, T. K. et al. Pyruvate kinase M2 is a poor prognostic marker of and a therapeutic target in ovarian cancer. PLoS ONE 12, e0182166 (2017).
Kalir, T. et al. Immunohistochemical staining of GLUT1 in benign, borderline, and malignant ovarian epithelia. Cancer 94, 1078–1082 (2002).
Rai, G. et al. Discovery and optimization of potent, cell-active pyrazole-based inhibitors of lactate dehydrogenase (LDH). J. Med. Chem. 60, 9184–9204 (2017).
Fantin, V. R., St-Pierre, J. & Leder, P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 (2006).
Nayak, A. P., Kapur, A., Barroilhet, L. & Patankar, M. S. Oxidative Phosphorylation: A Target for Novel Therapeutic Strategies Against Ovarian Cancer. Cancers (Basel). 10, 337 (2018).
Thuwajit, C., Ferraresi, A., Titone, R., Thuwajit, P. & Isidoro, C. The metabolic cross-talk between epithelial cancer cells and stromal fibroblasts in ovarian cancer progression: Autophagy plays a role. Med Res Rev. 38, 1235–1254 (2018).
Zhong, X. et al. Complex metabolic interactions between ovary, plasma, urine, and hair in ovarian cancer. Front Oncol. 12, 916375 (2022).
Fong, M. Y., McDunn, J. & Kakar, S. S. Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS ONE 6, e19963 (2011).
Maddocks, O. D. et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546 (2013).
Ying, H. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012).
Son, J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013).
Koundouros, N. et al. Metabolic fingerprinting links oncogenic PIK3CA with enhanced arachidonic acid-derived eicosanoids. Cell 181, 1596-1611 e1527 (2020).
Wu, S. et al. Targeting glutamine dependence through GLS1 inhibition suppresses ARID1A-inactivated clear cell ovarian carcinoma. Nat Cancer. 2, 189–200 (2021).
Anadon, C. M. et al. Ovarian cancer immunogenicity is governed by a narrow subset of progenitor tissue-resident memory T cells. Cancer Cell 40, 545-557 e513 (2022).
Jimenez-Sanchez, A. et al. heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell 170, 927-938 e920 (2017).
Laskowski, T. J., Biederstadt, A. & Rezvani, K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat. Rev. Cancer 22, 557–575 (2022).
Page, A., Chuvin, N., Valladeau-Guilemond, J. & Depil, S. Development of NK cell-based cancer immunotherapies through receptor engineering. Cell. Mol. Immunol. 21, 315–331 (2024).
Gonzalez, V. D. et al. High-grade serous ovarian tumor cells modulate NK cell function to create an immune-tolerant microenvironment. Cell Rep. 36, 109632 (2021).
Chiossone, L., Dumas, P. Y., Vienne, M. & Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18, 671–688 (2018).
Morvan, M. G. & Lanier, L. L. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 16, 7–19 (2016).
Opitz, C. A. et al. The therapeutic potential of targeting tryptophan catabolism in cancer. Br. J. Cancer 122, 30–44 (2020).
Prendergast, G. C., Malachowski, W. P., DuHadaway, J. B. & Muller, A. J. Discovery of IDO1 inhibitors: From bench to bedside. Cancer Res. 77, 6795–6811 (2017).
Liu, M. et al. Targeting the IDO1 pathway in cancer: From bench to bedside. J. Hematol. Oncol. 11, 100 (2018).
Munn, D. H., Sharma, M. D., Johnson, T. S. & Rodriguez, P. IDO, PTEN-expressing Tregs and control of antigen-presentation in the murine tumor microenvironment. Cancer Immunol. Immunother. 66, 1049–1058 (2017).
Guo, Y. et al. Indoleamine 2,3-dioxygenase (Ido) inhibitors and their nanomedicines for cancer immunotherapy. Biomaterials 276, 121018 (2021).
Liu, X. et al. Selective inhibition of IDO1 effectively regulates mediators of antitumor immunity. Blood 115, 3520–3530 (2010).
Della Chiesa, M. et al. The tryptophan catabolite L-kynurenine inhibits the surface expression of NKp46- and NKG2D-activating receptors and regulates NK-cell function. Blood 108, 4118–4125 (2006).
Stiff, A. et al. Nitric oxide production by myeloid-derived suppressor cells plays a role in impairing Fc receptor-mediated natural killer cell function. Clin. Cancer Res. 24, 1891–1904 (2018).
Geiger, R. et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829-842 e813 (2016).
Peyraud, F. et al. Circulating L-arginine predicts the survival of cancer patients treated with immune checkpoint inhibitors. Ann Oncol. 33, 1041–1051 (2022).
Pang, Z. et al. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
Sunami, K. et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 110, 1480–1490 (2019).
Kanke, Y. et al. Gene aberration profile of tumors of adolescent and young adult females. Oncotarget 9, 6228–6237 (2018).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Nagasaki, M. et al. Rare variant discovery by deep whole-genome sequencing of 1070 Japanese individuals. Nat. Commun. 6, 8018 (2015).
Robinson, J. T., Thorvaldsdottir, H., Wenger, A. M., Zehir, A. & Mesirov, J. P. Variant Review with the integrative genomics viewer. Cancer Res. 77, e31–e34 (2017).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Acknowledgements
The authors thank Hitoshi Ichikawa, Maiko Matsuda, Yoko Shimada, Sachiyo Mitani, Miyu Narita, and other physicians and staff members at the National Cancer Center Hospital for assistance and support. We also thank Bioedit Ltd. for assisting with English language editing.
Funding
This work was supported by Japan Agency for Medical Research and Development (AMED) (23ama221520h0001 to K.S.), a Grant-in-Aid for Scientific Research (B) 20H03668, BRIDGE (programs for bridging the gap between R&D and the ideal society (Society 5.0 to RH and KS) and generating economic and social value to K.S.), the National Cancer Center Research and Development Fund (2022-A-20, 2023-J-2, NCC Biobank, and NCC Core Facility to K.S.), and the Yamagata Prefectural Government and City of Tsuruoka (HM).
Author information
Authors and Affiliations
Contributions
Conceptualization, D. H., E. F., M. K-Kato, K. H., Y. A., M. K., R. H., K. M., A. S., Y. T., A. I., K. T., K. T., H. Y., and M. I.; methodology, M. Y. and K.S.; validation, M. Y. and K.S.; formal analysis, H. M. and H. O.; investigation, M. Y. and H. M.; resources, T.K, H. T, and M. I; data curation, H. O.; writing—original draft preparation, M. Y. and K. S.; writing—review and editing, K. S.; visualization, M. Y.; project administration, D. H., E. F., M. K-Kato, K. H., Y. A., M. K., R. H., K. M., A. S., Y. T., A. I., K. T., K. T., H. Y., and M. I.; funding acquisition K. S. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study was approved by the Institutional Review Board of the National Cancer Center Research Institute (2015-159). We confirmed that all methods were carried out in accordance with relevant guidelines and regulations.
Informed consent
All subjects and/or their guardians provided informed consent to participate in the study.
Registry and the registration no. of the study/trial
Not applicable.
Animal studies
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yamaguchi, M., Higuchi, D., Yoshida, H. et al. Metabolomic and transcriptomic analyses identify metabolic alterations and immune suppression in ovarian cancer. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38014-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38014-8