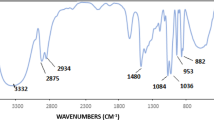
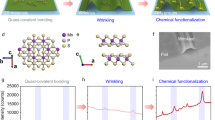
Abstract
For the safety monitoring of herbal medicines (HMs), HM inspectors carry out an organoleptic examination before acceptance for market supply. The organoleptic test processes are often regarded as labor-intensive, thus calling for efficient and reliable aids. Here, we propose a plasmonic artificial HM inspector based on a collaboration between surface-enhanced Raman spectroscopy (SERS) and deep learning (DL). Inherently, a SERS spectrum of an HM specimen contains several peaks that match bioactive compounds in the sample, yielding so-called fingerprint information of HM. Besides, its rapid, few-second data-acquisition speed lends the SERS-DL analysis adaptability to a complementary inspection method for organoleptic examination. Regarding the accuracy and reliability of this new method, the synergistic integration of SERS with DL attains ~95% accuracy in labor-saving differentiation of 35 HM species with similar appearances or of the same genus. Our SERS-DL-based analysis can potentially aid the organoleptic HM inspection and help upgrade the HM database, along with images and other analytical chemistry data.
Similar content being viewed by others
Data availability
Data available on request from the authors.
Code availability
Code availability on request from the authors.
References
Petrovska, B. B. Historical review of medicinal plants’ usage. Pharmacogn Rev. 6, 1–5 (2012).
Yin, S. Y., Wei, W. C., Jian, F. Y. & Yang, N. S. Therapeutic applications of herbal medicines for cancer patients. Evid. Based Complement Altern. Med. 2013, 302426 (2013).
Safarzadeh, E., Sandoghchian Shotorbani, S. & Baradaran, B. Herbal medicine as inducers of apoptosis in cancer treatment. Adv. Pharm. Bull. 4, 421–427 (2014).
Stickel, F. & Schuppan, D. Herbal medicine in the treatment of liver diseases. Dig. Liver Dis. 39, 293–304 (2007).
Li, X. Z., Zhang, S. N., Liu, S. M. & Lu, F. Recent advances in herbal medicines treating Parkinson’s disease. Fitoterapia 84, 273–285 (2013).
Gan, X. et al. Network medicine framework reveals generic herb-symptom effectiveness of traditional Chinese medicine. Sci. Adv. 9, eadh0215 (2023).
Park, J. & Choi, G. Review on herbal medicinal materials in the Korean pharmacopoeia and the Korean herbal pharmacopoeia. Korean Herb. Med. Inf. 4, 9–35 (2016).
The Dispensatory on the Visual and Organoleptic Examination of Herbal Medicine. (National Institute of Food and Drug Safety Evaluation, 2022).
Wagner, H. & Bladt, S. Plant Drug Analysis: A Thin Layer Chromatography Atlas (Springer, 1996).
Kumar, S., Jyotirmayee, K. & Sarangi, M. Thin layer chromatography: A tool of biotechnology for isolation of bioactive compounds from medicinal plants. Int. J. Pharm. Sci. Rev. Res. 18, 126–132 (2013).
EMA/HMPC/CHMP/CVMP. Guideline on specifications: Test procedures and acceptance criteria for herbal substances, herbal preparations and herbal medicinal products/traditional herbal medicinal products. In París Obtenido de Www Ema Europa Eu (2011).
Zeng, Z. D. et al. Mass spectral profiling: An effective tool for quality control of herbal medicines. Anal. Chim. Acta. 604, 89–98 (2007).
Zhang, A., Sun, H. & Wang, X. Mass spectrometry-driven drug discovery for development of herbal medicine. Mass Spectrom. Rev. 37, 307–320 (2018).
Komakech, R. et al. GC-MS and LC-TOF-MS profiles, toxicity, and macrophage-dependent in vitro anti-osteoporosis activity of Prunus africana (Hook f.) Kalkman bark. Sci. Rep. 12, 7044 (2022).
Jin, Z. et al. Intelligent SERS navigation system guiding brain tumor surgery by intraoperatively delineating the metabolic acidosis. Adv. Sci. (Weinh). 9, e2104935 (2022).
Eisemann, N. et al. Nationwide real-world implementation of AI for cancer detection in population-based mammography screening. Nat. Med. 31, 917–924 (2025).
Chang, Y. W. et al. Artificial intelligence for breast cancer screening in mammography (AI-STREAM): Preliminary analysis of a prospective multicenter cohort study. Nat. Commun. 16, 2248 (2025).
Shin, H. J., Han, K., Ryu, L. & Kim, E. K. The impact of artificial intelligence on the reading times of radiologists for chest radiographs. Npj Digit. Med. 6, 82 (2023).
Fan, X., Ming, W., Zeng, H., Zhang, Z. & Lu, H. Deep learning-based component identification for the Raman spectra of mixtures. Analyst 144, 1789–1798 (2019).
Chen, S. N. et al. Raman spectroscopy reveals abnormal changes in the urine composition of prostate cancer: An application of an intelligent diagnostic model with a deep learning algorithm. Adv. Intell. Syst. 3, 2000090 (2021).
Thrift, W. J. et al. Deep learning analysis of vibrational spectra of bacterial lysate for rapid antimicrobial susceptibility testing. ACS Nano. 14, 15336–15348 (2020).
Rho, E. et al. Separation-free bacterial identification in arbitrary media via deep neural network-based SERS analysis. Biosens. Bioelectron. 202, 113991 (2022).
Ji, K. X. et al. A deep learning strategy for discrimination and detection of multi-sulfonamides residues in aquatic environments using gold nanoparticles-decorated violet phosphorene SERS substrates. Sens. Actuators B-Chemical. 386, 133736 (2023).
Kim, S. et al. Plasma exosome analysis for protein mutation identification using a combination of Raman spectroscopy and deep learning. ACS Sens. 8, 2391–2400 (2023).
Fang, G. et al. Porous nanoframe based plasmonic structure with high-density hotspots for the quantitative detection of gaseous benzaldehyde. Small 21, 2408670 (2025).
Wang, X. et al. Design of highly sensitive plasmonic nanocavities and the application in trace detection of chromium (VI) ion. Anal. Chem. 97, 28068–28078 (2025).
Wang, X. et al. Plasmonic nanocavities with metallic spacer layers for ultrasensitive detection of Cr6 + Ions. ACS Photonics. 12, 3754–3762 (2025).
Lawrence, S., Giles, C. L., Tsoi, A. C. & Back, A. D. Face recognition: A convolutional neural-network approach. IEEE Trans. Neural Netw. 8, 98–113 (1997).
Zhou, B. et al. A 10 million image database for scene recognition. IEEE Trans. Pattern Anal. Mach. Intell. 40, 1452–1464 (2018).
Collobert, R. et al. Natural language processing (almost) from scratch. J. Mach. Learn. Res. 12, 2493–2537 (2011).
Ho, C. S. et al. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 10, 4927 (2019).
Lu, W., Chen, X., Wang, L., Li, H. & Fu, Y. V. Combination of an artificial intelligence approach and laser tweezers Raman spectroscopy for microbial identification. Anal. Chem. 92, 6288–6296 (2020).
Fan, W., Lu, X., Wang, J. & He, Y. Nanotechnologies for traditional Chinese medicine research: Quality control and therapeutic applications. Nanoscale 17, 27151–27171 (2025).
Xu, C. X., Song, P., Yu, Z. & Wang, Y. H. Surface-enhanced Raman spectroscopy as a powerful method for the analysis of Chinese herbal medicines. Analyst 149, 46–58 (2024).
Shi, G. et al. Fabrication of multifunctional SERS platform based on Ag NPs self-assembly ag-AAO nanoarray for direct determination of pesticide residues and baicalein in real samples. Coatings 11, 1054 (2021).
Wang, S. et al. Carbon dot- and gold nanocluster-based three-channel fluorescence array sensor: visual detection of multiple metal ions in complex samples. Sens. Actuators B. 369, 132194 (2022).
Song, L. et al. Highly sensitive SERS detection for aflatoxin B1 and Ochratoxin A based on aptamer-functionalized photonic crystal microsphere array. Sens. Actuators B. 364, 131778 (2022).
Liu, Q. H. et al. Rapid discrimination between wild and cultivated ophiocordyceps sinensis through comparative analysis of label-free SERS technique and mass spectrometry. Curr. Res. Food Sci. 9, 100820 (2024).
Du, M. et al. A new method for rapid identification of traditional Chinese medicine based on a new silver sol: using the SERS spectrum for quality control of flavonoids and flavonoid glycosides in potentilla discolor Bge. J. Mater. Chem. C. 11, 15138–15148 (2023).
Lin, H. et al. Surface-enhanced Raman on gold nanoparticles for the identification of the most common adulterant of astragali radix. Spectrosc. Lett. 51, 389–394 (2018).
Xiao, M. et al. Predicting the storage time of green tea by myricetin based on surface-enhanced Raman spectroscopy. Npj Sci. Food. 7, 28 (2023).
Zhang, H. et al. Surface-enhanced Raman scattering spectra revealing the inter-cultivar differences for Chinese ornamental Flos chrysanthemum: A new promising method for plant taxonomy. Plant. Methods. 13, 92 (2017).
Jiang, S. et al. Advanced SERSome-based artificial-intelligence technology for identifying medicinal and edible homologs. Talanta 292, 127931 (2025).
Sun, X. et al. Deep learning-enabled mobile application for efficient and robust herb image recognition. Sci. Rep. 12, 6579 (2022).
Lv, B. et al. Traditional Chinese medicine recognition based on target detection. Evid.-Based Complem. Altern. Med. 2022, 9220443 (2022).
Zhang, Y. et al. TCMP-300: A comprehensive traditional Chinese medicinal plant dataset for plant recognition. Sci. Data. 12, 1166 (2025).
Sasidharan, S., Chen, Y., Saravanan, D., Sundram, K. M. & Yoga Latha, L. Extraction, isolation and characterization of bioactive compounds from plants’ extracts. Afr. J. Tradit Complement. Altern. Med. 8, 1–10 (2011).
Han, Z. et al. Molecular origin of the Raman signal from Aspergillus nidulans conidia and observation of fluorescence vibrational structure at room temperature. Sci. Rep. 10, 5428 (2020).
Abubakar, A. R. & Haque, M. Preparation of medicinal plants: Basic extraction and fractionation procedures for experimental purposes. J. Pharm. Bioallied Sci. 12, 1–10 (2020).
Altun, A. O., Youn, S. K., Yazdani, N., Bond, T. & Park, H. G. Metal-dielectric-CNT nanowires for femtomolar chemical detection by surface enhanced Raman spectroscopy. Adv. Mater. 25, 4431–4436 (2013).
Altun, A. O., Bond, T., Pronk, W. & Park, H. G. Sensitive detection of competitive molecular adsorption by surface-enhanced Raman spectroscopy. Langmuir 33, 6999–7006 (2017).
Zou, Q., Huang, Y., Zhang, W., Lu, C. & Yuan, J. A comprehensive review of the pharmacology, chemistry, traditional uses and quality control of star anise (Illicium verum Hook. F.): An aromatic medicinal plant. Molecules 28, 7378 (2023).
Spiro, T. G. & Stein, P. Resonance effects in vibrational scattering from complex molecules. Annu. Rev. Phys. Chem. 28, 501–521 (1977).
Wiley, J. Sons, Inc. SpectraBase. (2023).
Rønsted, N. et al. Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol. Biol. 12, 1–12 (2012).
Hao, D. C. & Xiao, P. G. Pharmaceutical resource discovery from traditional medicinal plants: Pharmacophylogeny and pharmacophylogenomics. Chin. Herb. Med. 12, 104–117 (2020).
Liu, J. et al. Deep convolutional neural networks for Raman spectrum recognition: A unified solution. Analyst 142, 4067–4074 (2017).
Cheng, H. T. et al. ACM International Conference Proceeding Series. 7–10 (2016).
Arplt, D. et al. 34th International Conference on Machine Learning, ICML 2017. 350–359 (2017).
Sun, X. & Qian, H. Chinese herbal medicine image recognition and retrieval by convolutional neural network. PLOS ONE. 11, e0156327 (2016).
Cho, J. et al. Identification of toxic herbs using deep learning with focus on the sinomenium Acutum, aristolochiae manshuriensis Caulis, Akebiae Caulis. Appl. Sci. 9, 5456 (2019).
Miao, J., Huang, Y., Wang, Z., Wu, Z. & Lv, J. Image recognition of traditional Chinese medicine based on deep learning. Front. Bioeng. Biotechnol. 11, 1199803 (2023).
Weng, J. C., Hu, M. C. & Lan, K. C. Proceedings of the 8th ACM Multimedia Systems Conference, MMSys 2017. 233–234 (2017).
Tian, D., Zhou, C., Wang, Y., Zhang, R. & Yao, Y. NB-TCM-CHM: Image dataset of the Chinese herbal medicine fruits and its application in classification through deep learning. Data Brief 110405 (2024).
Hao, W., Han, M., Yang, H., Hao, F. & Li, F. A novel Chinese herbal medicine classification approach based on EfficientNet. Syst. Sci. Control Eng. 9, 304–313 (2021).
Hao, W., Han, M., Li, S. & Li, F. MTAL: A novel Chinese herbal medicine classification approach with mutual triplet attention learning. Wirel. Commun. Mobile Comput. 2022, 8034435 (2022).
Han, M., Zhang, J., Zeng, Y., Hao, F. & Ren, Y. A novel method of Chinese herbal medicine classification based on mutual learning. Mathematics 10, 1557 (2022).
Fei, C. et al. Identification of the raw and processed crataegi fructus based on the electronic nose coupled with chemometric methods. Sci. Rep. 11, 1849 (2021).
Wang, Y. et al. An optimized deep convolutional neural network for dendrobium classification based on electronic nose. Sens. Actuators A: Phys. 307, 111874 (2020).
Yang, S. et al. A novel method for rapid discrimination of bulbus of fritillaria by using electronic nose and electronic tongue technology. Anal. Methods. 7, 943–952 (2015).
Guerra-Nuñez, C. et al. The nucleation, radial growth, and bonding of TiO2 deposited via atomic layer deposition on single-walled carbon nanotubes. Appl. Surf. Sci. 555 (2021).
Sun, Y. et al. Highly sensitive surface-enhanced Raman scattering substrate made from superaligned carbon nanotubes. Nano Lett. 10, 1747–1753 (2010).
Park, S. G. et al. 3D hybrid plasmonic nanomaterials for highly efficient optical absorbers and sensors. Adv. Mater. 27, 4290–4295 (2015).
Baek, S. J., Park, A., Ahn, Y. J. & Choo, J. Baseline correction using asymmetrically reweighted penalized least squares smoothing. Analyst 140, 250–257 (2015).
Savitzky, A. & Golay, M. J. E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964).
He, K., Zhang, X., Ren, S. & Sun, J. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 770–778 (2016).
Funding
This work was funded by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (No. 2020R1A3B2079741). A part of this work was supported by the Po-Ca Networking Groups funded by the Postech-Catholic Biomedical Engineering Institute (PCBMI) (No. 2.0080459.01).
Author information
Authors and Affiliations
Contributions
H.K. contributed conceptualization, methodology, investigation, and writing - original draft; J.L. contributed software, data curation, writing - original draft, and visualization; S.W.K. contributed conceptualization; H.G.P. contributed conceptualization, writing - review & editing, and supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, H., Lee, J., Kim, S.W. et al. Plasmonic artificial inspector for herbal medicines via surface-enhanced Raman spectroscopy and deep learning. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38497-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38497-5