Abstract
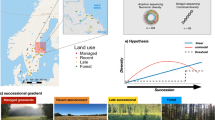
Soil microorganisms are essential for sustaining ecosystem functions, driving biogeochemical cycles, and modulating carbon storage. However, the nutrient-mediated mechanisms by which different land-use types shape soil microbial communities remain unclear. This study investigated three typical land-use types—plantation, grassland, and high-standard cropland—in Xinyang City, China, to evaluate their effects on soil microbial community. Results showed that soil nutrient contents—including total nitrogen, total phosphorus, alkaline-hydrolyzable nitrogen, and available phosphorus—as well as microbial alpha diversity indices, were consistently higher in topsoil than in subsoil and more pronounced in plantation than in grassland and cropland. Acidobacteriota, Pseudomonadota, Ascomycota, and Basidiomycota dominated across all land uses, though community composition varied significantly among them. Network analysis revealed strongest microbial connectivity in plantation, intermediate in grassland, and weakest in cropland. Our findings demonstrate that land-use type and soil depth directly affect soil available nutrients, thereby influencing microbial diversity. This study clarifies the nutrient-driven pathways through which land use affects soil ecosystems, providing important insights for sustainable land management and ecological conservation.
Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2025) in National Genomics Data Center (Nucleic Acids Res 2025), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA033082; CRA033083) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
References
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E. & van der Heijden, M. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10, 4841. https://doi.org/10.1038/s41467-019-12798-y (2019).
Delgado-Baquerizo, M. et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat. Ecol. Evol. 4, 210–220. https://doi.org/10.1038/s41559-019-1084-y (2020).
Chen, Y. et al. Conversion of natural grassland to cropland alters microbial community assembly across northern China. Environ. Microbiol. 24, 5630–5642. https://doi.org/10.1111/1462-2920.16127 (2022).
Han, Z. et al. Microbial diversity and the abundance of keystone species drive the response of soil multifunctionality to organic substitution and biochar amendment in a tea plantation. GCB Bioenergy. 14, 481–495. https://doi.org/10.1111/gcbb.12926 (2022).
Delgado-Baquerizo, M. et al. Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 20, 1295–1305. https://doi.org/10.1111/ele.12826 (2017).
Delgado-Baquerizo, M. et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 10541. https://doi.org/10.1038/ncomms10541 (2016).
Li, J. et al. Plant productivity and microbial composition drive soil carbon and nitrogen sequestrations following cropland abandonment. Sci. Total Environ. 744, 140802. https://doi.org/10.1016/j.scitotenv.2020.140802 (2020).
Kong, Y. L. et al. Research progress on the mechanism by which soil microorganisms affect soil health. Acta Pedol. Sin. 61, 331–347 (2024).
Zhu, R. et al. Response of carbohydrate-degrading enzymes and microorganisms to land use change in the southeastern Qinghai-Tibetan Plateau,China. Appl. Soil. Ecol. 200, 105442. https://doi.org/10.1016/j.apsoil.2024.105442 (2024).
Chen, Y., Ma, S., Jiang, H., Hu, Y. & Lu, X. Influences of litter diversity and soil moisture on soil microbial communities in decomposing mixed litter of alpine steppe species. Geoderma 377, 114577. https://doi.org/10.1016/j.geoderma.2020.114577 (2020).
Goss-Souza, D. et al. Soil microbial community dynamics and assembly under long-term land use change. Fems Microbiol. Ecol. 93, fix109. https://doi.org/10.1093/femsec/fix109 (2017).
Huang, J. et al. Changes of soil bacterial community, network structure, and carbon, nitrogen and sulfur functional genes under different land use types. Catena 231, 107385. https://doi.org/10.1016/j.catena.2023.107385 (2023).
de Vries, F. T. et al. Soil food web properties explain ecosystem services across European land use systems. P Natl. Acad. Sci. Usa. 110, 14296–14301. https://doi.org/10.1073/pnas.1305198110 (2013).
Gomez, E., Garland, J. & Conti, M. Reproducibility in the response of soil bacterial community-level physiological profiles from a land use intensification gradient. Appl. Soil. Ecol. 26, 21–30. https://doi.org/10.1016/j.apsoil.2003.10.007 (2004).
Wang, Q. et al. Conversion of wetlands to farmland and forests reduces soil microbial functional diversity and carbon use intensity. Appl. Ecol. Env Res. 20, 4553–4564. https://doi.org/10.15666/aeer/2005_45534564 (2022).
Guan, Z. et al. Soil microbial communities response to different fertilization regimes in young Catalpa bungei plantation. Front. Microbiol. 13, 948875. https://doi.org/10.3389/fmicb.2022.948875 (2022).
Qiao, L., Guan, Z., Ren, F. & Ma, T. Comparative analysis of rhizosphere microbial communities in monoculture and mixed oak-pine forests: Structural and functional insights. Front. Microbiol. 16, 1646535. https://doi.org/10.3389/fmicb.2025.1646535 (2025).
Lu, R. Agricultural and Chemistry Analysis of Soil (China Agricultural Science and Technology, 2002).
Callahan, B. J. et al. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods. 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. https://doi.org/10.1186/s40168-018-0470-z (2018).
Vazquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2, 16. https://doi.org/10.1186/2047-217X-2-16 (2013).
Zhi, J. et al. Impact of land use patterns on the structure and function of soil bacterial communities. Bull. Bot. Res. 45, 22–33 (2025).
Seabloom, E. W., Borer, E. T. & Tilman, D. Grassland ecosystem recovery after soil disturbance depends on nutrient supply rate. Ecol. Lett. 23, 1756–1765. https://doi.org/10.1111/ele.13591 (2020).
Schilling, E. M., Waring, B. G., Schilling, J. S. & Powers, J. S. Forest composition modifies litter dynamics and decomposition in regenerating tropical dry forest. Oecologia 182, 287–297. https://doi.org/10.1007/s00442-016-3662-x (2016).
Gao, H., Gong, J., Ye, T., Maier, M. & Liu, J. Constructing cropland ecological stability assessment method based on disturbance-resistance-response processes and classifying cropland ecological types. Sci. Total Environ. 930, 172673. https://doi.org/10.1016/j.scitotenv.2024.172673 (2024).
Zhang, M. et al. Land use intensification alters the relative contributions of plant functional diversity and soil properties on grassland productivity. Oecologia 201, 119–127. https://doi.org/10.1007/s00442-022-05288-4 (2023).
Qin, Y. et al. Changes of total phenols and condensed tannins during the decomposition of mixed leaf litter of Pinus massoniana and broad-leaved trees. Chin. J. Appl. Ecol. 29, 2224–2232. https://doi.org/10.13287/j.1001-9332.201807.038 (2018).
Russo, T. A., Tully, K., Palm, C. & Neill, C. Leaching losses from Kenyan maize cropland receiving different rates of nitrogen fertilizer. Nutr. Cycl. Agroecosys. 108, 195–209. https://doi.org/10.1007/s10705-017-9852-z (2017).
Gao, G. et al. Effects of land-use patterns on soil carbon and nitrogen variations along revegetated hillslopes in the Chinese Loess Plateau. Sci. Total Environ. 746, 141156. https://doi.org/10.1016/j.scitotenv.2020.141156 (2020).
Niu, Y. et al. Variations in seasonal and inter-annual carbon fluxes in a semi-arid sandy maize cropland ecosystem in China’s Horqin Sandy Land. Environ. Sci. Pollut R. 29, 5295–5312. https://doi.org/10.1007/s11356-021-15751-z (2022).
Shukla, A. K., Behera, S. K., Lakaria, B. L. & Tripathi, A. Effect of land use and soil depth on the distribution of phyto-available nutrients and SOC pools of vertisols in central India. Environ. Monit. Assess. 195, 1405. https://doi.org/10.1007/s10661-023-12032-9 (2023).
Gabriel, C. E., Kellman, L. & Prest, D. Examining mineral-associated soil organic matter pools through depth in harvested forest soil profiles. Plos One. 13, e206847. https://doi.org/10.1371/journal.pone.0206847 (2018).
Yan, K. et al. Depth-dependent patterns of soil microbial community in the E-waste dismantling area. J. Hazard. Mater. 444, 130379. https://doi.org/10.1016/j.jhazmat.2022.130379 (2023).
Xiao, E., Wang, Y., Xiao, T., Sun, W. & Ning, Z. Microbial community responses to land-use types and its ecological roles in mining area. Sci. Total Environ. 775, 145753. https://doi.org/10.1016/j.scitotenv.2021.145753 (2021).
He, D. et al. Microbial life-history strategies and genomic traits between pristine and cropland soils. Msystems 10, e17825. https://doi.org/10.1128/msystems.00178-25 (2025).
Llado, S., Lopez-Mondejar, R. & Baldrian, P. Drivers of microbial community structure in forest soils. Appl. Microbiol. Biot. 102, 4331–4338. https://doi.org/10.1007/s00253-018-8950-4 (2018).
Zhao, M. et al. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant. Cell. Environ. 44, 613–628. https://doi.org/10.1111/pce.13928 (2021).
Liu, L. et al. Medicinal fungi and soil: interactions, plant interactions, and ecological restoration for sustainable use: A review. Int. J. Med. Mushrooms. 27, 61–74. https://doi.org/10.1615/IntJMedMushrooms.2025059592 (2025).
Yang, Y. et al. Responses of soil microbial diversity, network complexity and multifunctionality to three land-use changes. Sci. Total Environ. 859, 160255. https://doi.org/10.1016/j.scitotenv.2022.160255 (2023).
Guo, J. et al. Linkages between plant community composition and soil microbial diversity in Masson Pine forests. Plants-Basel 12, 1750. https://doi.org/10.3390/plants12091750 (2023).
Hannula, S. E. et al. Time after time: Temporal variation in the effects of grass and forb species on soil bacterial and fungal communities. Mbio 10, e02635–e02619. https://doi.org/10.1128/mBio.02635-19 (2019).
Li, B. et al. Stoichiometric imbalances between soil microorganisms and their resources regulate litter decomposition. Funct. Ecol. 37, 3136–3149. https://doi.org/10.1111/1365-2435.14459 (2023).
Karimi, B. et al. Microbial diversity and ecological networks as indicators of environmental quality. Environ. Chem. Lett. 15, 265–281. https://doi.org/10.1007/s10311-017-0614-6 (2017).
Banerjee, S. et al. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. Isme J. 13, 1722–1736. https://doi.org/10.1038/s41396-019-0383-2 (2019).
Zhang, J. et al. Soil microbial richness predicts ecosystem multifunctionality through co-occurrence network complexity in alpine meadow. Acta Ecol. Sin. 42, 2542–2558 (2022).
Barberan, A., Bates, S. T., Casamayor, E. O. & Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. Isme J. 6, 343–351. https://doi.org/10.1038/ismej.2011.119 (2012).
Zheng, J., Shi, J. & Wang, D. Diversity of soil fungi and entomopathogenic fungi in subtropical mountain forest in Southwest China. Env Microbiol. Rep. 16, e13267. https://doi.org/10.1111/1758-2229.13267 (2024).
Xin, Y., Wu, Y., Zhang, H., Li, X. & Qu, X. Soil depth exerts a stronger impact on microbial communities and the sulfur biological cycle than salinity in salinized soils. Sci. Total Environ. 894, 164898. https://doi.org/10.1016/j.scitotenv.2023.164898 (2023).
Acknowledgements
We thank this work was supported by the National Key Wildlife Conservation Project (GTH [2024]153). We would like to express our special gratitude to Professors Chunxiao Song and Tianxiao Ma for their review and revision of the manuscript.
Funding
This study was supported by the National Key Wildlife Conservation Project (GTH [2024]153).
Author information
Authors and Affiliations
Contributions
GH, YR and XH conceived the idea; GH, RY, XH and SH performed the experiments, data analysis, prepared the figures and wrote the manuscript; ZG, CS and TM edited the manuscript, and all authors gave approval for the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, G., Rong, Y., Song, C. et al. Influence of land-use types on soil microbial communities and nutrient changes in Xinyang City, China. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38635-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38635-z