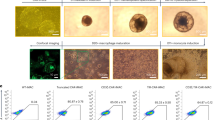
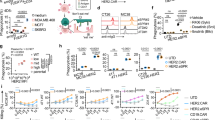
Abstract
Chimeric antigen receptor macrophages (CAR-M) are emerging as a next-generation cellular modality for therapies ranging from viral infection to solid tumors, leveraging innate phagocytic and antigen-presenting functions. Here, we compared CAR constructs incorporating intracellular signaling domains (ICDs) derived from CD3ζ, Fc gamma receptor IIa (CD32a), complement receptor 3 (CR3), and Toll-like receptor 4 (TLR4) in THP-1-derived monocytes and macrophages. Using an anti-viral SARS-CoV-2 model as a screening platform, we subsequently validated key findings in an anti-tumor mesothelin (MSLN) model. Results indicated that CARCD32a exhibited superior phagocytic capacity compared with CARCD3ζ in both monocytes and macrophages. While combining CR3 (CD11b and CD18) and CD32a domains did not enhance phagocytosis, it significantly increased the expression of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α). The incorporation of TLR4 signaling domain reduced surface CAR expression and phagocytic capacity but markedly increased inflammatory cytokine induction, suggesting that TLR4-driven cytokine production can be enhanced despite diminished phagocytosis in this setting. Furthermore, following phagocytosis, CAR-monocytes induced antigen-specific CD8+ T cell activation via antigen presentation. Collectively, these findings highlight CD32a-based and combinatorial ICD designs as a framework for functionally tuned CAR-M platform for solid tumor immunotherapy and anti-viral applications.
Similar content being viewed by others
Data availability
All data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Pan, K. et al. CAR race to cancer immunotherapy: from CAR T, CAR NK to CAR macrophage therapy. J. Exp. Clin. Cancer Res. 41, 119 (2022).
Hou, Y. et al. Beyond CAR-T cells: exploring CAR-NK, CAR-M, and CAR-γδ T strategies in solid tumor immunotherapy. Front. Immunol. 16, 1675807 (2025).
Chen, Y. et al. Developing CAR-immune cell therapy against SARS-CoV-2: current status, challenges and prospects. Biochem. Pharmacol. 222, 116066 (2024).
Wu, D. et al. CAR T-cell therapy in autoimmune diseases: a promising frontier on the horizon. Front. Immunol. 16, 1613878 (2025).
Gao, J. et al. Efficacy and safety of allogeneic CD19 CAR NK-cell therapy in systemic lupus erythematosus: a case series in China. Lancet 406, 2968–2979 (2025).
Cao, Q. et al. Targeting inflammation with chimeric antigen receptor macrophages using a signal switch. Nat. Biomed. Eng. 9, 1502–1516 (2025).
Li, J., Liu, C., Zhang, P., Shen, L. & Qi, C. Optimizing CAR T cell therapy for solid tumours: a clinical perspective. Nat. Rev. Clin. Oncol. 22, 953–968 (2025).
Li, T. et al. CAR-NK cells for cancer immunotherapy: recent advances and future directions. Front. Immunol. 15, 1361194 (2024).
Alrehaili, M., Couto, P. S., Khalife, R. & Rafiq, Q. A. CAR-macrophages in solid tumors: promise, progress, and prospects. Npj Precis Onc. 9, 382 (2025).
Khan, A. N., Asija, S., Pendhari, J. & Purwar, R. CAR-T cell therapy in hematological malignancies: where are we now and where are we heading for? Eur. J. Haematol. 112, 6–18 (2024).
Zhang, X., Zhu, L., Zhang, H., Chen, S. & Xiao, Y. CAR-T cell therapy in hematological malignancies: current opportunities and challenges. Front. Immunol. 13, 927153 (2022).
Shi, Y. et al. Exploring the potential of CAR-macrophage therapy. Life Sci. 361, 123300 (2025).
Fu, W. et al. CAR macrophages for SARS-CoV-2 immunotherapy. Front Immunol 12, (2021).
Lei, A. et al. A second-generation M1-polarized CAR macrophage with antitumor efficacy. Nat. Immunol. 25, 102–116 (2024).
Zhang, H. et al. Silencing of SIRPα enhances the antitumor efficacy of CAR-M in solid tumors. Cell. Mol. Immunol. 21, 1335–1349 (2024).
Freeman, S. A. & Grinstein, S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol. Rev. 262, 193–215 (2014).
Chen, S. et al. Macrophages in immunoregulation and therapeutics. Sig Transduct. Target. Ther. 8, 207 (2023).
Mantovani, A., Allavena, P., Marchesi, F. & Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 21, 799–820 (2022).
Muntjewerff, E. M. & Meesters, L. D. & van den Bogaart, G. Antigen Cross-Presentation by macrophages. Front Immunol 11, (2020).
Klichinsky, M. et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 38, 947–953 (2020).
Gao, Z. et al. Targeting cardiac fibrosis with chimeric antigen receptor macrophages. Cell. Discov. 10, 86 (2024).
Wang, J. et al. CAR-Macrophage therapy alleviates myocardial Ischemia-Reperfusion injury. Circul. Res. 135, 1161–1174 (2024).
Pierini, S. et al. Chimeric antigen receptor macrophages (CAR-M) sensitize HER2 + solid tumors to PD1 Blockade in pre-clinical models. Nat. Commun. 16, 706 (2025).
Reiss, K. A. et al. CAR-macrophage therapy for HER2-overexpressing advanced solid tumors: a phase 1 trial. Nat. Med. 31, 1171–1182 (2025).
Huang, T., Bei, C., Hu, Z. & Li, Y. CAR-macrophage: breaking new ground in cellular immunotherapy. Front. Cell. Dev. Biol. 12, 1464218 (2024).
Liu, M. et al. CAR-Macrophages and CAR-T cells synergistically kill tumor cells in vitro. Cells 11, 3692 (2022).
Hadiloo, K., Taremi, S., Heidari, M. & Esmaeilzadeh, A. The CAR macrophage cells, a novel generation of chimeric antigen-based approach against solid tumors. Biomark. Res. 11, 103 (2023).
Kloosterman, D. J. & Akkari, L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell 186, 1627–1651 (2023).
Duan, Z. & Luo, Y. Targeting macrophages in cancer immunotherapy. Signal. Transduct. Target. Ther. 6, 127 (2021).
Duan, Z., Li, Z., Wang, Z., Chen, C. & Luo, Y. Chimeric antigen receptor macrophages activated through TLR4 or IFN-γ receptors suppress breast cancer growth by targeting VEGFR2. Cancer Immunol. Immunother. 72, 3243–3257 (2023).
Walbaum, S. et al. Complement receptor 3 mediates both sinking phagocytosis and phagocytic cup formation via distinct mechanisms. J. Biol. Chem. 296, 100256 (2021).
McInturff, J. E., Modlin, R. L. & Kim, J. The role of Toll-like receptors in the pathogenesis and treatment of dermatological disease. J. Invest. Dermatology. 125, 1–8 (2005).
Sharma, V., Sharma, P. & Singh, T. G. Mechanistic insights on TLR-4 mediated inflammatory pathway in neurodegenerative diseases. Pharmacol. Rep. 76, 679–692 (2024).
Clevers, H., Alarcon, B., Wileman, T. & Terhorst, C. The T cell receptor/CD3 complex: a dynamic protein ensemble. Annu. Rev. Immunol. 6, 629–662 (1988).
Lan, J. et al. Structure of the SARS-CoV-2 Spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Bettini, M. L. et al. Cutting edge: CD3 ITAM diversity is required for optimal TCR signaling and thymocyte development. J. Immunol. 199, 1555–1560 (2017).
Anania, J. C., Chenoweth, A. M., Wines, B. D. & Hogarth, P. M. The human FcγRII (CD32) family of leukocyte FcR in health and disease. Front Immunol 10, (2019).
Hickman, E. et al. Expanded characterization of in vitro polarized M0, M1, and M2 human monocyte-derived macrophages: bioenergetic and secreted mediator profiles. PLOS ONE. 18, e0279037 (2023).
Suleimanov, S. K. et al. Radical-Generating Activity, Phagocytosis, and mechanical properties of four phenotypes of human macrophages. Int. J. Mol. Sci. 25, 1860 (2024).
Affandi, A. J. et al. CD169 defines activated CD14 + Monocytes with enhanced CD8 + T cell activation capacity. Front Immunol 12, (2021).
Brossart, P. The role of antigen spreading in the efficacy of immunotherapies. Clin. Cancer Res. 26, 4442–4447 (2020).
Gulley, J. L. et al. Role of antigen spread and distinctive characteristics of immunotherapy in cancer treatment. JNCI: J. Natl. Cancer Inst. 109, djw261 (2017).
Liang, Y., Xu, Q. & Gao, Q. Advancing CAR-based immunotherapies in solid tumors: CAR- macrophages and neutrophils. Front. Immunol. 14, 1291619 (2023).
Lu, J. et al. CAR macrophages: a promising novel immunotherapy for solid tumors and beyond. Biomark. Res. 12, 86 (2024).
Saha, S. et al. High expression of mesothelin in plasma and tissue is associated with poor prognosis and promotes invasion and metastasis in gastric cancer. Adv. Cancer Biology - Metastasis. 7, 100098 (2023).
Lv, J. & Li, P. Mesothelin as a biomarker for targeted therapy. Biomark. Res. 7, 18 (2019).
Faust, J. R., Hamill, D., Kolb, E. A., Gopalakrishnapillai, A. & Barwe, S. P. Mesothelin: an immunotherapeutic target beyond solid tumors. Cancers 14, 1550 (2022).
Chung, J. Y. et al. Rapid CAR Screening and circRNA-driven CAR-NK Cells for Persistent Shed-resistant Immunotherapy. 08.19.671003 Preprint at (2025). https://doi.org/10.1101/2025.08.19.671003 (2025).
Chowdhury, P. S., Viner, J. L., Beers, R. & Pastan, I. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Proceedings of the National Academy of Sciences 95, 669–674 (1998).
Chowdhury, P. S. & Pastan, I. Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat. Biotechnol. 17, 568–572 (1999).
Lucas, M., Zhang, X., Prasanna, V. & Mosser, D. M. ERK activation following macrophage FcγR ligation leads to chromatin modifications at the IL-10 Locus1. J. Immunol. 175, 469–477 (2005).
Li, J., Chen, P. & Ma, W. The next frontier in immunotherapy: potential and challenges of CAR-macrophages. Experimental Hematol. Oncol. 13, 76 (2024).
Morva, A., Arroyo, A. B., Andreeva, L. & Tapia-Abellán, A. Luengo-Gil, G. Unleashing the power of CAR-M therapy in solid tumors: a comprehensive review. Front. Immunol. 16, 1615760 (2025).
Huo, Y. et al. M1 polarization enhances the antitumor activity of chimeric antigen receptor macrophages in solid tumors. J. Transl Med. 21, 225 (2023).
Zhang, W. et al. Macrophage polarization in the tumor microenvironment: emerging roles and therapeutic potentials. Biomed. Pharmacother. 177, 116930 (2024).
Sterner, R. C. & Sterner, R. M. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 11, 69 (2021).
Feucht, J. et al. Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat. Med. 25, 82–88 (2019).
Ciesielska, A., Matyjek, M. & Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 78, 1233–1261 (2021).
Wang, P. et al. Chimeric antigen receptor with novel intracellular modules improves antitumor performance of T cells. Signal. Transduct. Target. Ther. 10, 20 (2025).
Genin, M., Clement, F., Fattaccioli, A., Raes, M. & Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to Etoposide. BMC Cancer. 15, 577 (2015).
Lee, H. N. et al. Novel FRET-based immunological synapse biosensor for the prediction of chimeric antigen Receptor-T cell function. Small Methods. 9, 2401016 (2025).
Kang, C. K. et al. Broad humoral and cellular immunity elicited by one-dose mRNA vaccination 18 months after SARS-CoV-2 infection. BMC Med. 20, 181 (2022).
He, J. et al. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69, 6705–6711 (1995).
Connor, R. I., Chen, B. K., Choe, S. & Landau, N. R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206, 935–944 (1995).
Kang, C. K. et al. Longitudinal analysis of human memory T-Cell response according to the severity of illness up to 8 months after severe acute respiratory syndrome coronavirus 2 infection. J. Infect. Dis. 224, 39–48 (2021).
Kang, C. K. et al. Distinct immune response at 1 year Post-COVID-19 according to disease severity. Front. Immunol. 13, 830433 (2022).
Acknowledgements
English grammar was reviewed and revised by ChatGPT, a language model developed by OpenAI in San Francisco, CA, USA.
Funding
This work was supported in part by the Creative-Pioneering Researchers Program through Seoul National University (to C.-H. L. and H.-R. Kim) and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIT) (RS-2021-NR056559 to H.-R. Kim, RS-2024-00440679 to C.-H.L.).
Author information
Authors and Affiliations
Contributions
C.-H.L. and J.S.H. conceived and designed the project. J.S.H., S.J.L., and Y.J.K. performed the experiments. C.-H.L., H.R.K., and J.S.H. analyzed the data. C.-H.L., H.R.K., J.S.H., C.K.K., W.B.P., and H.M.S. wrote the manuscript. All authors have reviewed the manuscript and approved its submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hong, J., Lee, S., Kim, Y. et al. Optimization of THP-1-CAR monocytes utilizing CD32a signaling phagocytosis for antigen-specific T cell activation. Sci Rep (2026). https://doi.org/10.1038/s41598-026-39406-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-39406-6