Abstract
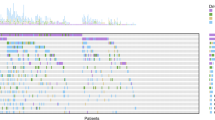
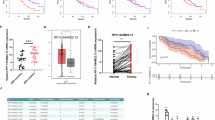
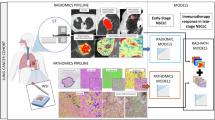
This study aimed to identify a novel plasma lncRNA biomarker and establish a model based on lncRNAs and radiomics for predicting the response of non-small cell lung cancer (NSCLC) to chemo- and radio-therapy. Next-generation sequencing and integrated bioinformatics analysis were used to identify lncRNAs associated with the response of NSCLC to chemo- and radio-therapy. RT-qPCR was utilized to detect MIF-AS1 expression in the plasma of NSCLC patients. Radiomics analysis was performed on CT images of NSCLC patients. The model was constructed by multiple logistic regression. The expression of the lncRNA MIF-AS1 was up-regulated in the plasma of patients with chemo- and radio-resistant NSCLC, as validated by RT-qPCR in 124 NSCLC patients. Furthermore, in vitro experiments demonstrated that knockdown of MIF-AS1 expression significantly reduced cellular proliferation and invasion, and increased the sensitivity of NSCLC cells to the chemotherapeutic drug cisplatin. Using the ceRNA network, a DNA-damage repair related protein RAD21 was identified as a target gene of MIF-AS1. Finally, two radiomic features were found to be associated with the response of NSCLC to chemo- and radio-therapy. Combining the MIF-AS1 level and the two radiomic features, a model was established to predict the response of NSCLC to chemo- and radio-therapy, with a high AUC of 0.808. MIF-AS1 could be a novel biomarker for predicting the response of NSCLC to chemo- and radio-therapy. This model, which uses both CT radiomics and MIF-AS1 levels, increases the accuracy of predicting therapeutic response in NSCLC patients.
Similar content being viewed by others
Data availability
The raw sequence data of exosomal lncRNAs reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA014950) that are publicly accessible at https:/ngdc.cncb.ac.cn/gsa-human.
Code availability
Not applicable.
References
Chen, Y., Zitello, E., Guo, R. & Deng, Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin. Transl. Med. 11, e367. https://doi.org/10.1002/ctm2.367 (2021).
Lin, Y., Leng, Q., Zhan, M. & Jiang, F. A Plasma Long Noncoding RNA Signature for Early Detection of Lung Cancer. Translational oncology 11, 1225–1231. https://doi.org/10.1016/j.tranon.2018.07.016 (2018).
Deans, A. J. & West, S. C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 11, 467–480. https://doi.org/10.1038/nrc3088 (2011).
Chen, Z. et al. Long non-coding RNA in lung cancer. Clinica chimica acta Int. J. Clin. Chem. 504, 190–200. https://doi.org/10.1016/j.cca.2019.11.031 (2020).
Liu, H. Y. et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J. Investigative Med. : Official Publication of the Am. Federation for Clin. Res. 68, 52–59. https://doi.org/10.1136/jim-2019-001080 (2020).
Liu, G. et al. Expression and significance of LncRNA MNX1-AS1 in non-small cell lung cancer. Onco. Targets. Ther. 12, 3129–3138. https://doi.org/10.2147/ott.S198014 (2019).
Zhang, X. et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 19, 47. https://doi.org/10.1186/s12943-020-01171-z (2020).
Guiducci, G. & Stojic, L. Long Noncoding RNAs at the Crossroads of Cell Cycle and Genome Integrity. Trends in genetics : TIG 37, 528–546. https://doi.org/10.1016/j.tig.2021.01.006 (2021).
Sun, W., Zu, Y., Fu, X. & Deng, Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol. Rep. 38, 3347–3354. https://doi.org/10.3892/or.2017.6056 (2017).
Zhang, M. et al. Long Noncoding RNA CRNDE/PRC2 Participated in the Radiotherapy Resistance of Human Lung Adenocarcinoma Through Targeting p21 Expression. Oncol. Res. 26, 1245–1255. https://doi.org/10.3727/096504017x14944585873668 (2018).
Liu, L. et al. Long non-coding RNA ANRIL promotes homologous recombination-mediated DNA repair by maintaining ATR protein stability to enhance cancer resistance. Mol. Cancer 20, 94. https://doi.org/10.1186/s12943-021-01382-y (2021).
Tamang, S. et al. SNHG12: An LncRNA as a Potential Therapeutic Target and Biomarker for Human Cancer. Front. Oncol. 9, 901. https://doi.org/10.3389/fonc.2019.00901 (2019).
Li, C. et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell. Physiol. 234, 20721–20727. https://doi.org/10.1002/jcp.28678 (2019).
Nagasaka, M. et al. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol. Cancer 20, 82. https://doi.org/10.1186/s12943-021-01371-1 (2021).
Lin, L. Y. et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer 17, 84. https://doi.org/10.1186/s12943-018-0834-9 (2018).
Min, L. et al. Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int. J. Clin. Oncol. 27, 1013–1024. https://doi.org/10.1007/s10147-022-02129-5 (2022).
Cucchiara, F. et al. Combining liquid biopsy and radiomics for personalized treatment of lung cancer patients State of the art and new perspectives. Pharmacological Res. 169, 105643. https://doi.org/10.1016/j.phrs.2021.105643 (2021).
Wen, Q., Yang, Z., Dai, H., Feng, A. & Li, Q. Radiomics Study for Predicting the Expression of PD-L1 and Tumor Mutation Burden in Non-Small Cell Lung Cancer Based on CT Images and Clinicopathological Features. Front. Oncol. 11, 620246. https://doi.org/10.3389/fonc.2021.620246 (2021).
He, B., Dong, D., She, Y., Zhou, C., Fang, M., Zhu, Y., Zhang, H., Huang, Z., Jiang, T., Tian, J.; et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. Journal for immunotherapy of cancer 2020, 8, https://doi.org/10.1136/jitc-2020-000550.
Wang, J. et al. MiR-320b/RAD21 axis affects hepatocellular carcinoma radiosensitivity to ionizing radiation treatment through DNA damage repair signaling. Cancer Sci. 112, 575–588. https://doi.org/10.1111/cas.14751 (2021).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Holdenrieder, S. et al. Carcinoembryonic antigen and cytokeratin-19 fragments for assessment of therapy response in non-small cell lung cancer: a systematic review and meta-analysis. Br. J. Cancer 116, 1037–1045. https://doi.org/10.1038/bjc.2017.45 (2017).
Fan, Y. et al. LncRNA MIF-AS1 aggravates the progression of ovarian cancer by sponging miRNA-31-5p. Eur. Rev. Med. Pharmacol. Sci. 24, 2248–2255. https://doi.org/10.26355/eurrev_202003_20490 (2020).
Ni, P. et al. Correlation of MIF-AS1 polymorphisms with the risk and prognosis of gastric cancer. Pathol. Res. Pract. 233, 153850. https://doi.org/10.1016/j.prp.2022.153850 (2022).
Ding, J., Wu, W., Yang, J. & Wu, M. Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249-3p/HOXB8 axis. Pathol. Res. Pract. 215, 152376. https://doi.org/10.1016/j.prp.2019.03.005 (2019).
Nasmyth, K. & Haering, C. H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558. https://doi.org/10.1146/annurev-genet-102108-134233 (2009).
Nishiyama, T. Cohesion and cohesin-dependent chromatin organization. Curr. Opin. Cell Biol. 58, 8–14. https://doi.org/10.1016/j.ceb.2018.11.006 (2019).
Perea-Resa, C., Wattendorf, L., Marzouk, S. & Blower, M. D. Cohesin: behind dynamic genome topology and gene expression reprogramming. Trends Cell Biol. 31, 760–773. https://doi.org/10.1016/j.tcb.2021.03.005 (2021).
Kuru-Schors, M., Haemmerle, M. & Gutschner, T. The Cohesin Complex and Its Interplay with Non-Coding RNAs. Non-coding RNA 2021, 7, https://doi.org/10.3390/ncrna7040067.
van Agthoven, T. et al. Selective recruitment of breast cancer anti-estrogen resistance genes and relevance for breast cancer progression and tamoxifen therapy response. Endocr. Relat. Cancer 17, 215–230. https://doi.org/10.1677/erc-09-0062 (2010).
Mendes-Pereira, A. M. et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc. Natl. Acad. Sci. U.S.A. 109, 2730–2735. https://doi.org/10.1073/pnas.1018872108 (2012).
Atienza, J. M. et al. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 4, 361–368. https://doi.org/10.1158/1535-7163.Mct-04-0241 (2005).
Zhao, J. et al. The regulatory and predictive functions of miR-17 and miR-92 families on cisplatin resistance of non-small cell lung cancer. BMC Cancer 15, 731. https://doi.org/10.1186/s12885-015-1713-z (2015).
Trebeschi, S. et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Annals Oncology : official J. Eur. Soc. Med. Oncology 30, 998–1004. https://doi.org/10.1093/annonc/mdz108 (2019).
Li, J., Ge, S., Sang, S., Hu, C. & Deng, S. Evaluation of PD-L1 Expression Level in Patients With Non-Small Cell Lung Cancer by (18)F-FDG PET/CT Radiomics and Clinicopathological Characteristics. Front. Oncol. 11, 789014. https://doi.org/10.3389/fonc.2021.789014 (2021).
Wang, J. et al. CT radiomics-based model for predicting TMB and immunotherapy response in non-small cell lung cancer. BMC Med. Imaging 24, 45. https://doi.org/10.1186/s12880-024-01221-8 (2024).
Lin, Y. et al. A classifier integrating plasma biomarkers and radiological characteristics for distinguishing malignant from benign pulmonary nodules. Int. J. Cancer 141, 1240–1248. https://doi.org/10.1002/ijc.30822 (2017).
Xing, W. et al. A prediction model based on DNA methylation biomarkers and radiological characteristics for identifying malignant from benign pulmonary nodules. BMC Cancer 21, 263. https://doi.org/10.1186/s12885-021-08002-4 (2021).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81872438), the Program of Research and Development of Key Common Technologies and Engineering of Major Scientific and Technological Achievements in Hefei (2021YL007), the Collaborative Innovation Program of Hefei Science Center, CAS (2022HSC-CIP015), and the Program of Clinical Medical Translational Research in Anhui Province (202304295107020092).
Funding
This study was supported by the National Natural Science Foundation of China (81872438), the Program of Research and Development of Key Common Technologies and Engineering of Major Scientific and Technological Achievements in Hefei (2021YL007), the Collaborative Innovation Program of Hefei Science Center, CAS (2022HSC-CIP015), and the Program of Clinical Medical Translational Research in Anhui Province (202304295107020092).
Author information
Authors and Affiliations
Contributions
HW and BH designed the study. FY, JY, YY, ZH, HJ, YZ (Yucheng Zhang) and YZ (Yani Zhang) collected the samples and performed the in vitro experiments. FY, JW (Jiexiao Wang), JW (Jialiang Wang), QZ and JQ performed the bioinformatics analysis. FY and JZ wrote the first draft of the manuscript. BH and HW revised the manuscript. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by The Ethics Committee of Hefei Cancer Hospital of the Chinese Academy of Sciences, with the approval number SL-PJ2023-098.
Consent to participate
All clinical samples and information were obtained with written informed consent of the patients.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ye, F., Yin, Y., Wang, J. et al. A lncRNA and radiomics-based model for predicting the response of non-small cell lung cancer to chemo- and radio-therapy. Sci Rep (2026). https://doi.org/10.1038/s41598-026-39560-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-39560-x