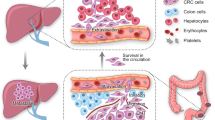
Abstract
Nearly half of colorectal cancer patients develop liver metastases. While surgical removal offers a potential cure, the majority experience recurrence within two years. Accurate tools to predict the recurrence risk are lacking. This study proposes a multi-omics framework combining computed tomography, transcriptomic (RNA) sequencing, and the Clinical Risk Score (CRS) to predict the likelihood of two-year recurrence after colorectal liver metastasis (CRLM) resection. Our approach addresses undetected RNA transcripts by introducing generative adversarial imputation and leverages generative learning and transformers to manage high dimensional gene expression data. Imaging features are extracted using a foundation model alongside interpretable radiomics. Tested on a prospectively maintained dataset of 129 patients, the pipeline achieved an area under the curve of 0.75 ± 0.05, outperforming unimodal and bimodal approaches and the CRS. This assistive tool can improve risk stratification, inform patients of their expected outcomes, guide follow-up care and inspire clinical trials for post-operative treatments.
Similar content being viewed by others
Data availability
Anonymized imaging, RNA sequencing, and associated patient clinicopathological characteristics are available upon reasonable request from the study authors, conditional on approval by our Research Ethics Board. The underlying code to implement the proposed pipeline will be made publicly available on https://github.com/rksaber/recurrence_prediction.
Code availability
The underlying code to implement the proposed pipeline will be made publicly available on https://github.com/rksaber/recurrence_prediction.
References
Siegel, R. L. et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164 (2020).
Leung, U. et al. Colorectal cancer liver metastases and concurrent extrahepatic disease treated with resection. Ann. Surg. 265, 158–165 (2017).
Amoretti, M. et al. Production and detection of cold antihydrogen atoms. Nature 419, 456–459 (2002).
Buisman, F. E. et al. Recurrence patterns after resection of colorectal liver metastasis are modified by perioperative systemic chemotherapy. World J. Surg. 44, 876–886 (2020).
Tomlinson, J. S. et al. Actual 10-year survival after resection of colorectal liver metastases defines cure. J. Clin. Oncol. 25, 4575–4580 (2007).
Fong, Y., Fortner, J., Sun, R. L., Brennan, M. F. & Blumgart, L. H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann. Surg. 230, 309–18; discussion 318–21 (1999).
Wong, G. Y. M. et al. Performance of prognostic models incorporating KRAS mutation status to predict survival after resection of colorectal liver metastases. HPB 24, 1316–1325 (2022).
Teng, S. et al. Tissue-specific transcription reprogramming promotes liver metastasis of colorectal cancer. Cell Res. 30, 34–49 (2019).
Li, L., Sun, M., Wang, J. & Wan, S. Multi-omics based artificial intelligence for cancer research. In Cutting Edge Artificial Intelligence Spatial Transcriptomics and Proteomics Approaches Analyze Cancer, Vol. 163 of Advances in Cancer Research, (eds Madan, E., Fisher, P. B. & Gogna, R.) Ch. 9 303–356 (Academic Press, 2024).
Wu, B. et al. Prognosis prediction of stage IV colorectal cancer patients by mRNA transcriptional profile. Cancer Med. 11, 4900–4912 (2022).
Staal, F. C. et al. Radiomics for the prediction of treatment outcome and survival in patients with colorectal cancer: a systematic review. Clin. Colorectal Cancer 20, 52–71 (2021).
Montagnon, E. et al. Radiomics analysis of baseline computed tomography to predict oncological outcomes in patients treated for resectable colorectal cancer liver metastasis. PLoS ONE 19, e0307815 (2024).
Lubner, M. G. et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom. Imaging 40, 2331–2337 (2015).
Ye, S. et al. Association of CT-based delta radiomics biomarker with progression-free survival in patients with colorectal liver metastases undergo chemotherapy. Front. Oncol. 12, 843991 (2022).
Aerts, H. J. W. L. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Zhang, S. & Metaxas, D. On the challenges and perspectives of foundation models for medical image analysis. Med. Image Anal. 91, 102996 (2024).
Wesdorp, N. et al. Advanced image analytics predicting clinical outcomes in patients with colorectal liver metastases: a systematic review of the literature. Surg. Oncol. 38, 101578 (2021).
Maaref, A. et al. Predicting the response to FOLFOX-based chemotherapy regimen from untreated liver metastases on baseline CT: a deep neural network approach. J. Digit. Imaging 33, 937–945 (2020).
Granata, V. et al. Radiomics and machine learning analysis by computed tomography and magnetic resonance imaging in colorectal liver metastases prognostic assessment. Radiol. Med. 128, 1310–1332 (2023).
Luo, X. et al. Prognostication of colorectal cancer liver metastasis by CE-based radiomics and machine learning. Transl. Oncol. 47, 101997 (2024).
Keyl, J. et al. Deep learning-based assessment of body composition and liver tumour burden for survival modelling in advanced colorectal cancer. J. Cachexia Sarcopenia Muscle 14, 545–552 (2023).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Wang, Q. et al. Exploring tumor heterogeneity in colorectal liver metastases by imaging: unsupervised machine learning of preoperative ct radiomics features for prognostic stratification. Eur. J. Radiol. 175, 111459 (2024).
Yang, M. et al. A multi-omics machine learning framework in predicting the survival of colorectal cancer patients. Comput. Biol. Med. 146, 105516 (2022).
Lu, W. et al. Folfox treatment response prediction in metastatic or recurrent colorectal cancer patients via machine learning algorithms. Cancer Med. 9, 1419–1429 (2020).
Sun Tian, S. L., Fulong, W. & Chen, G. Identification of two subgroups of folfox resistance patterns and prediction of folfox response in colorectal cancer patients. Cancer Investig. 39, 62–72 (2021).
Amniouel, S. & Jafri, M. S. High-accuracy prediction of colorectal cancer chemotherapy efficacy using machine learning applied to gene expression data. Front. Physiol. 14, 1272206 (2024).
Kim, J. et al. Transcriptomes of the tumor-adjacent normal tissues are more informative than tumors in predicting recurrence in colorectal cancer patients. J. Transl. Med. 21, 209 (2023).
Zheng, S. et al. Machine learning-based screening and validation of liver metastasis-specific genes in colorectal cancer. Sci. Rep. 14, 17679 (2024).
Alleman, K. et al. Multimodal deep learning-based prognostication in glioma patients: a systematic review. Cancers 15, 545 (2023).
Ho, D., Tan, I. B. H. & Motani, M. Predictive models for colorectal cancer recurrence using multi-modal healthcare data. In Proc. Conference Health, Inference, and Learning, CHIL ’21, 204–213 (Association for Computing Machinery, 2021).
Chaudhary, K., Poirion, O. B., Lu, L. & Garmire, L. X. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 24, 1248–1259 (2017).
Pink, R. C. et al. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 17, 792–798 (2011).
Zajac, P. et al. MAGE-A antigens and cancer immunotherapy. Front. Med. 4, 18 (2017).
Weon, J. L. & Potts, P. R. The MAGE protein family and cancer. Curr. Opin. Cell Biol. 37, 1–8 (2015).
Lu, T. et al. Adamts18 deficiency promotes colon carcinogenesis by enhancing β-catenin and p38MAPK/ERK1/2 signaling in the mouse model of AOM/DSS-induced colitis-associated colorectal cancer. Oncotarget 8, 18979–18990 (2017).
Zhang, X. et al. Loss of CHGA protein as a potential biomarker for colon cancer diagnosis: a study on biomarker discovery by machine learning and confirmation by immunohistochemistry in colorectal cancer tissue microarrays. Cancers 14, 2664 (2022).
Wang, M., Miao, Z., Cen, H., He, J. & Wei, C. Long non-coding RNA (LncRNA) CHROMR promotes the expression of the CNNM1 gene by adsorbing hsa-mir-1299 to obtain drug resistance in diffuse large B lymphoma cells. Transl. Cancer Res. 11, 1362–1371 (2022).
Kim, S.-L. et al. Lipocalin 2 negatively regulates cell proliferation and epithelial to mesenchymal transition through changing metabolic gene expression in colorectal cancer. Cancer Sci. 108, 2176–2186 (2017).
Carion, N. et al. End-to-End Object Detection with Transformers. In Computer Vision – ECCV 2020. ECCV 2020. Lecture Notes in Computer Science, (eds Vedaldi, A., Bischof, H., Brox, T., Frahm, JM.) vol. 12346, https://doi.org/10.1007/978-3-030-58452-8_13 (Springer, Cham, 2020).
Kirillov, A. et al. Segment Anything. In IEEE/CVF International Conference on Computer Vision (ICCV), 3992-4003, https://doi.org/10.1109/ICCV51070.2023.00371 (Paris, France, 2023).
Radford, A. et al. Learning Transferable Visual Models From Natural Language Supervision. In Proceedings of the 38th International Conference on Machine Learning. In Proceedings of Machine Learning Research 139:8748-8763, Available from https://proceedings.mlr.press/v139/radford21a.html.
Girdhar, R. et al. Imagebind: one embedding space to bind them all. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 15180-15190 (2023).
Ma, J. et al. Segment anything in medical images. Nat. Commun. 15, 654 (2024).
Thawakar, O. et al. XrayGPT: Chest Radiographs Summarization using Large Medical Vision-Language Models. In Proceedings of the 23rd Workshop on Biomedical Natural Language Processing, 440–448 (Bangkok, Thailand, 2024).
Pai, S. et al. Foundation model for cancer imaging biomarkers. Nat. Mach. Intell. 6, 354–367 (2024).
Saber, R. et al. Radiomics using computed tomography to predict CD73 expression and prognosis of colorectal cancer liver metastases. J. Transl. Med. 21, 507 (2023).
Wesdorp, N. et al. Identifying genetic mutation status in patients with colorectal cancer liver metastases using radiomics-based machine-learning models. Cancers 15, 5648 (2023).
Elforaici, M. E. A. et al. Semi-supervised vit knowledge distillation network with style transfer normalization for colorectal liver metastases survival prediction. Med. Image Anal. 99, 103346 (2025).
Vorontsov, E., Tang, A., Pal, C. & Kadoury, S. Liver lesion segmentation informed by joint liver segmentation https://arxiv.org/abs/1707.07734 (2018).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol. 15, 550 (2014).
Zhang, Y., Parmigiani, G. & Johnson, W. E. Combat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinformatics 2, lqaa078 (2020).
Yoon, J., Jordon, J. & van der Schaar, M. GAIN: Missing data imputation using generative adversarial nets https://arxiv.org/abs/1806.02920 (2018).
Saber, R., Routy, B., Turcotte, S. & Kadoury, S. RNA sequencing-based histological subtyping of non-small cell lung cancer with generative adversarial data imputation. In Proc. IEEE-EMBS International Conference on Biomedical and Health Informatics 1–4 (IEEE, 2023).
Xu, L., Skoularidou, M., Cuesta-Infante, A. & Veeramachaneni, K. Modeling tabular data using conditional GAN. In Advances in Neural Information Processing Systems,Vol. 32 (2019).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77 21, e104–e107 (2017).
Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58, 267–288 (1996).
Chen, T. et al. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the 37th International Conference on Machine Learning, In Proceedings of Machine Learning Research 119:1597-1607. Available from https://proceedings.mlr.press/v119/chen20j.html (2020).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Wang, F. & Liu, H. Understanding the behaviour of contrastive loss. In Proc.Conference on Computer Vision and Pattern Recognition 2495–2504 (IEEE, 2021).
Bakr, S. et al. Data for NSCLC radiogenomics (version 4). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2017.7hs46erv Data set (2017).
Messaoudi, N. et al. Prognostic value of CD73 expression in resected colorectal cancer liver metastasis. Oncoimmunology 9, 1746138 (2020).
Vickers, A. J., van Calster, B. & Steyerberg, E. W. A simple, step-by-step guide to interpreting decision curve analysis. Diagn. Progn. Res. 3, 18 (2019).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. In Advance in Neural Information Processing Systems (eds Guyon, I. et al.) 4765–4774 (Curran Associates, Inc., 2017).
Arik, S. Ö. & Pfister, T. TabNet: Attentive Interpretable Tabular Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, 35, 6679-6687, https://doi.org/10.1609/aaai.v35i8.16826 (2021).
Huang, X., Khetan, A., Cvitkovic, M. & Karnin, Z. Tabtransformer: tabular data modeling using contextual embeddings https://arxiv.org/abs/2012.06678 (2020).
Hollmann, N., Müller, S., Eggensperger, K. & Hutter, F. TabPFN: a transformer that solves small tabular classification problems in a second. In Proc. 11th International Conference on Learning Representations (ICLR, 2023).
Acknowledgements
S.K., S.T., and A.T. are researchers at the Research Center of the University of Montreal Hospital (CRCHUM), supported by the Fonds de Recherche du Québec—Santé (FRQS). RS is supported by the Fonds de recherche du Québec—Nature et Technologie (FRQNT). ST is supported by the FRQS Clinician Scientist Senior Salary Award (No. 349577). ST obtained support for RNA sequencing by a Terry Fox Marathon of Hope Cancer Centres Network grant and pre-clinical research funding from Bristol Myers Squibb and Turnstone Biologics. The CRCHUM Hepatopancreatobiliary Cancer Biobank and Prospective Database is supported by the Université de Montréal Roger des Groseillers Research Chair in Hepatopancreatobiliary Surgical Oncology. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. The authors wish to express their gratitude to L. Rousseau, S. Langevin, and J. Bilodeau for their contributions to patient recruitment, biospecimen aquisition and processing, and to the CRCHUM histopathology core facility and R. Kosovskaia for RNA extraction and sequencing logistics. Moreover, M. Cerny, V. Hamilton, T. Derennes, and A. Ilinca are recognized for their work in editing segmentations and differentiating liver metastases from benign coexisting liver lesions.
Author information
Authors and Affiliations
Contributions
M.C. preprocessed RNA sequencing data. E.M. performed CT image collection and preparation. A.T. supervised the manual validation of CT segmentations. S.K. and S.T. performed project conceptualization and supervision tasks. R.S. implemented the methodology, performed formal analysis, data curation, software analysis, validation, visualization, and wrote the original draft. All authors reviewed the manuscript prior to submission.
Corresponding author
Ethics declarations
Competing interests
Outside the submitted work, S.T. has received consultant fees from Bristol Myers Squibb and Turnstone Biologics, speaking fees from Celgene and AstraZeneca, and has research funding from Lovance Biotherapeutics and Turnstone Biologics. The remaining authors have no financial or non-financial competing interests to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Saber, R., Carneiro, M., Montagnon, E. et al. Multi-omics fusion network for prediction of early recurrence in colorectal liver metastases. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-025-01264-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-025-01264-2