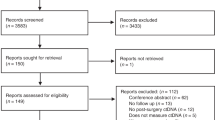
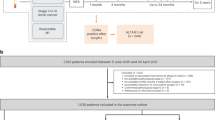
Abstract
Recurrence after curative-intent surgery for colorectal cancer is a major cause of cancer-related death. Circulating cell-free tumor DNA (ctDNA) is increasingly used in the perioperative setting to detect residual disease. However, the association between preoperative ctDNA, the tumor microenvironment, including tumor-infiltrating lymphocytes, and recurrence is unknown. We explored the association between ctDNA and the tumor microenvironment in patients with non-metastatic CRC undergoing curative-intent surgery. ctDNA was assessed using a tumor-agnostic hypermethylated cfDNA test. Among 140 patients, ctDNA tested positive in 102 (72.9%) before surgery, with 38 (27.1%) tumors classified as immune infiltration high. ctDNA was associated with expression of cancer-metastasis pathways, while immune active phenotypes were associated with immune infiltration high tumors. ctDNA status could identify deficient mismatch repair tumors with an active immune phenotype. The results suggest that a positive ctDNA analysis before surgery is associated with a metastatic tumor microenvironment.
Similar content being viewed by others
Data availability
Data is provided within the manuscript or supplementary data file. Code is available upon reasonable request to the corresponding author.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71, 209–249 (2021).
Kapiteijn, E. et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N. Engl. J. Med. 345, 638–646 (2001).
Bahadoer, R. R. et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): a randomised, open-label, phase 3 trial. Lancet Oncol. 22, 29–42 (2021).
Pagès, F. et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391, 2128–2139 (2018).
Bramsen, J. B. et al. Molecular-subtype-specific biomarkers improve prediction of prognosis in colorectal cancer. Cell Rep. 19, 1268–1280 (2017).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
El Sissy, C. et al. International validation of the immunoscore biopsy in patients with rectal cancer managed by a watch-and-wait strategy. J. Clin. Oncol. 42, 70–80 (2024).
Henriksen, T. V. et al. Circulating tumor DNA in stage III colorectal cancer, beyond minimal residual disease detection, toward assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin. Cancer Res. 28, 507–517 (2022).
Gögenur, M., Hadi, N. A. H., Qvortrup, C., Andersen, C. L. & Gögenur, I. ctDNA for risk of recurrence assessment in patients treated with neoadjuvant treatment: a systematic review and meta-analysis. Ann. Surg. Oncol. 29, 8666–8674 (2022).
Jensen, S. Ø. et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer- A clinical biomarker discovery and validation study. Clin. Epigenetics 11, 158 (2019).
Øgaard, N. et al. Tumour-agnostic circulating tumour DNA analysis for improved recurrence surveillance after resection of colorectal liver metastases: a prospective cohort study. Eur. J. Cancer 163, 163–176 (2022).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome Analytics. Cell 173, 400–416.e11 (2018).
Dongre, A. & Weinberg, R. A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019).
Ruan, K., Song, G. & Ouyang, G. Role of hypoxia in the hallmarks of human cancer. J. Cell Biochem. 107, 1053–1062 (2009).
Wu, Y. & Zhou, B. P. TNF-α/NF-κB/Snail pathway in cancer cell migration and invasion. Br. J. Cancer 102, 639–644 (2010).
Qian, D. C. et al. PI3K/Akt/mTOR signaling and plasma membrane proteins are implicated in responsiveness to adjuvant dendritic cell vaccination for metastatic colorectal cancer. Clin. Cancer Res. 23, 399–406 (2017).
Danaher, P. et al. Gene expression markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer 5, 18 (2017).
Louvi, A. & Artavanis-Tsakonas, S. Notch and disease: a growing field. Semin. Cell Dev. Biol. 23, 473–480 (2012).
Huang, F. & Chen, Y. G. Regulation of TGF-β receptor activity. Cell Biosci. 2, 9 (2012).
Fiehn, A. M. K., Reiss, B., Gögenur, M., Bzorek, M. & Gögenur, I. Development of a Fully automated method to obtain reproducible lymphocyte counts in patients with colorectal cancer. Appl. immunohistochem. Mol. morphology 30, 493–500 (2022).
Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 67, 301–320 (2005).
Moss J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nature Communications [Internet]. 9. Available from: https://doi.org/10.1038/s41467-018-07466-6 (2018).
Shirasawa, M. et al. Prognostic impact of peripheral blood neutrophil to lymphocyte ratio in advanced-stage pulmonary large cell neuroendocrine carcinoma and its association with the immune-related tumour microenvironment. Br. J. Cancer 124, 925–932 (2021).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224–ra24 (2014).
Forshew, T. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 4, 136–ra68 (2012).
Boysen A. K. et al. Cell-free DNA levels and correlation to stage and outcome following treatment of locally advanced rectal cancer. Tumor Biol. 39, 1010428317730976 (2017).
Anunobi, R. et al. Extracellular DNA promotes colorectal tumor cell survival after cytotoxic chemotherapy. J. Surg. Res. 226, 181–191 (2018).
Waldvogel Abramowski, S. et al. Cell-free nucleic acids are present in blood products and regulate genes of innate immune response. Transfusion 58, 1671–1681 (2018).
Korabecna, M. et al. Cell-free DNA in plasma as an essential immune system regulator. Sci. Rep. 10, 17478 (2020).
Grabuschnig, S. et al. Putative origins of cell-free DNA in humans: a review of active and passive nucleic acid release mechanisms. Int. J. Mol. Sci. 21, 1–24 (2020).
Gutierrez et al. Biomarker-directed, pembrolizumab-based combination therapy in non-small cell lung cancer: phase 2 KEYNOTE-495/KeyImPaCT trial interim results. Nat Med. 29, 1718–1727 (2023).
Jensen, S., Øgaard, N., Nielsen, H. J., Bramsen, J. B. & Andersen, C. L. Enhanced performance of DNA methylation markers by simultaneous measurement of sense and antisense DNA strands after cytosine conversion. Clin. Chem. 66, 925–933 (2020).
Dube, S., Qin, J. & Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE e2876, e2876 (2008).
Øgaard, N. et al. Circulating tumour DNA and risk of recurrence in patients with asymptomatic versus symptomatic colorectal cancer. Br. J. Cancer 131, 1707–1715 (2024).
Bhattacharya, A. et al. An approach for normalization and quality control for NanoString RNA expression data. Brief. Bioinforma. 22, bbaa163 (2021).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma. 32, 2847–2849 (2016).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics a J. Integr. Biol. 16, 284–287 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
Biorender.com and Adobe Illustrator was used for figure preparation. This study was funded by the Misse and Valdemar Risoms Foundation (IG, no grant number), the Toyota Foundation (IG, no grant number), the Novo Nordic Foundation (CLA grants no. NNF17OC0025052 and NNF22OC0074415), and the Danish Cancer Society (CLA grant no. R231-A13845).
Author information
Authors and Affiliations
Contributions
Conceptualization: M.G. and I.G. Methodology: M.G., I.G., and C.A. Investigation: M.G., S.J., N.Ø., I.L., M.B., R.D.B., and A.F. Visualization: M.G. and L.B. Funding acquisition: M.G., I.G., and C.L.A. Supervision: I.G. and C.A. Writing—original draft: M.G. Writing—review and editing: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gögenur, M., Balsevicius, L., Jensen, S.Ø. et al. Preoperative positive ctDNA analysis is associated with the tumor microenvironment, and the risk of recurrence in non-metastatic colorectal cancer. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-026-01288-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-026-01288-2