Abstract
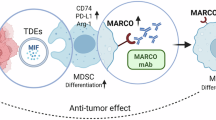
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest cancers worldwide. The role of macrophage receptor with collagenous structure (MARCO), a scavenger receptor class-A protein expressed on macrophage surface, in PDAC progression remains unclear. Here, we identified a subset of MARCO-expressing macrophages with strong immunosuppressive signatures that were markedly increased in PDAC patients. Analysis of MARCOhi PDAC samples displayed reduced proportion of CD8⁺ T cells and NK cells, accompanied by an increased proportion of regulatory T cells (Tregs). In vitro, co-culture with multiple PDAC cell lines potently induced MARCO expression on both human and murine macrophages, driving them to a pro-tumorigenic polarization phenotype. Cell-cell interaction analyses further indicated that vascular endothelial growth factor (VEGF) selectively targets MARCO⁺ macrophages, and VEGF stimulation significantly upregulates MARCO expression in vitro. Notably, genetic ablation of Marco markedly suppressed tumor growth in a murine PDAC model, at least partly through enhanced proportion of NK and T cells. Furthermore, MARCO⁺ macrophages were also enriched across several other cancer types, suggesting a potential pan-cancer relevance. Collectively, our findings uncover a critical role of MARCO⁺ macrophages in PDAC progression and highlight MARCO as a promising therapeutic target with potential applicability across multiple malignancies.
Similar content being viewed by others
Data availability
All data generated in the study are available in the present article, supplementary information, and supplementary data.
Code availability
The code supporting this study is not publicly available but will be made available by the corresponding author upon reasonable request.
References
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74, 12–49 (2024).
Calheiros, J. et al. A first-in-class inhibitor of homologous recombination DNA repair counteracts tumour growth, metastasis and therapeutic resistance in pancreatic cancer. J. Exp. Clin. Cancer Res 44, 129 (2025).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Wainberg, Z. A. et al. NALIRIFOX versus nab-paclitaxel and gemcitabine in treatment-naive patients with metastatic pancreatic ductal adenocarcinoma (NAPOLI 3): a randomised, open-label, phase 3 trial. Lancet 402, 1272–1281 (2023).
Zou, X. et al. Characterization of intratumoral tertiary lymphoid structures in pancreatic ductal adenocarcinoma: cellular properties and prognostic significance. J. Immunother. Cancer 11, e006698 (2023).
Bear, A. S., Vonderheide, R. H. & O’Hara, M. H. Challenges and opportunities for pancreatic cancer immunotherapy. Cancer Cell 38, 788–802 (2020).
Balachandran, V. P. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017).
Quaranta, V. et al. Macrophage-derived granulin drives resistance to immune checkpoint inhibition in metastatic pancreatic cancer. Cancer Res. 78, 4253–4269 (2018).
Zhou, J. et al. A novel role of TGFBI in macrophage polarization and macrophage-induced pancreatic cancer growth and therapeutic resistance. Cancer Lett. 578, 216457 (2023).
Bernard, V. et al. Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin. Cancer Res. 25, 2194–2205 (2019).
Caronni, N. et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature 623, 415–422 (2023).
Olaniru, O. E. et al. Single-cell transcriptomic and spatial landscapes of the developing human pancreas. Cell Metab. 35, 184–199.e5 (2023).
Li, W. et al. Identification and prognostic analysis of biomarkers to predict the progression of pancreatic cancer patients. Mol. Med. 28, 43 (2022).
Hosein, A. N. et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight 5, e129212 (2019).
Cui Zhou, D. et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nat. Genet. 54, 1390–1405 (2022).
Li, W., Wang, F., Guo, R., Bian, Z. & Song, Y. Targeting macrophages in hematological malignancies: recent advances and future directions. J. Hematol. Oncol. 15, 110 (2022).
Liu, M., Liu, L., Song, Y., Li, W. & Xu, L. Targeting macrophages: a novel treatment strategy in solid tumors. J. Transl. Med 20, 586 (2022).
Eisinger, S. et al. Targeting a scavenger receptor on tumor-associated macrophages activates tumor cell killing by natural killer cells. Proc. Natl. Acad. Sci. USA 117, 32005–32016 (2020).
Georgoudaki, A.-M. et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 15, 2000–2011 (2016).
La Fleur, L. et al. Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory T cells and supports cytotoxic lymphocyte function. Cancer Res. 81, 956–967 (2021).
Molgora, M. et al. IL-1R8 is a checkpoint in NK cells regulating anti-tumour and anti-viral activity. Nature 551, 110–114 (2017).
Sa, J. K. et al. Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol. 21, 216 (2020).
Shi, B. et al. The scavenger receptor MARCO expressed by tumor-associated macrophages are highly associated with poor pancreatic cancer prognosis. Front Oncol. 11, 771488 (2021).
Zhang, S. et al. Single cell transcriptomic analyses implicate an immunosuppressive tumor microenvironment in pancreatic cancer liver metastasis. Nat. Commun. 14, 5123 (2023).
Werba, G. et al. Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment. Nat. Commun. 14, 797 (2023).
Chen, K. et al. Immune profiling and prognostic model of pancreatic cancer using quantitative pathology and single-cell RNA sequencing. J. Transl. Med. 21, 210 (2023).
Chen, K. et al. Single-cell transcriptome profiling of primary tumors and paired organoids of pancreatobiliary cancer. Cancer Lett. 582, 216586 (2024).
Oh, K. et al. Coordinated single-cell tumor microenvironment dynamics reinforce pancreatic cancer subtype. Nat. Commun. 14, 5226 (2023).
Storrs, E. P. et al. High-dimensional deconstruction of pancreatic cancer identifies tumor microenvironmental and developmental stemness features that predict survival. NPJ Precis. Oncol. 7, 105 (2023).
Xu, L. et al. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res. 78, 6575–6580 (2018).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med 24, 1550–1558 (2018).
Song, G. et al. Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma. Nat. Commun. 13, 1642 (2022).
Zhang, J. et al. Single-cell transcriptome sequencing reveals aberrantly activated inter-tumor cell signaling pathways in the development of clear cell renal cell carcinoma. J. Transl. Med. 22, 37 (2024).
Chen, G. et al. Single-cell transcriptomic analysis reveals that the APP-CD74 axis promotes immunosuppression and progression of testicular tumors. J. Pathol. 264, 250–269 (2024).
Wheeler, K. C. et al. VEGF may contribute to macrophage recruitment and M2 polarization in the decidua. PLoS One 13, e0191040 (2018).
Wang, S. et al. Targeting M2-like tumor-associated macrophages is a potential therapeutic approach to overcome antitumor drug resistance. NPJ Precis. Oncol. 8, 31 (2024).
Zhou, W.-H. et al. The overexpression of fibronectin 1 promotes cancer progression and associated with M2 macrophages polarization in head and neck squamous cell carcinoma patients. Int. J. Gen. Med. 15, 5027–5042 (2022).
Gao, Y., Dai, W., Ouyang, Z., Shen, M. & Shi, X. Dendrimer-mediated intracellular delivery of fibronectin guides macrophage polarization to alleviate acute lung injury. Biomacromolecules 24, 886–895 (2023).
Matsubara, E. et al. The significance of SPP1 in lung cancers and its impact as a marker for protumor tumor-associated macrophages. Cancers 15, 2250 (2023).
Simons, M., Gordon, E. & Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17, 611–625 (2016).
Tchaikovski, V., Fellbrich, G. & Waltenberger, J. The molecular basis of VEGFR-1 signal transduction pathways in primary human monocytes. Arterioscler. Thromb. Vasc. Biol. 28, 322–328 (2008).
Lee, J. W., Komar, C. A., Bengsch, F., Graham, K. & Beatty, G. L. Genetically engineered mouse models of pancreatic cancer: the KPC model (LSL-Kras(G12D/+);LSL-Trp53(R172H/+);Pdx-1-Cre), its variants, and their application in immuno-oncology drug discovery. Curr. Protoc. Pharm. 73, 14.39.1–14.39.20 (2016).
Cassetta, L. & Pollard, J. W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887–904 (2018).
Ho, W. J., Jaffee, E. M. & Zheng, L. The tumour microenvironment in pancreatic cancer—clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 17, 527–540 (2020).
Griesmann, H. et al. Pharmacological macrophage inhibition decreases metastasis formation in a genetic model of pancreatic cancer. Gut 66, 1278–1285 (2017).
Nielsen, S. R. et al. Macrophage-secreted granulin supports pancreatic cancer metastasis by inducing liver fibrosis. Nat. Cell Biol. 18, 549–560 (2016).
Mass, E., Nimmerjahn, F., Kierdorf, K. & Schlitzer, A. Tissue-specific macrophages: how they develop and choreograph tissue biology. Nat. Rev. Immunol. 23, 563–579 (2023).
Nasir, I. et al. Tumor macrophage functional heterogeneity can inform the development of novel cancer therapies. Trends Immunol. 44, 971–985 (2023).
Revel, M., Sautès-Fridman, C., Fridman, W.-H. & Roumenina, L. T. C1q+ macrophages: passengers or drivers of cancer progression. Trends Cancer 8, 517–526 (2022).
Chen, K. et al. Single cell RNA-seq identifies immune-related prognostic model and key signature-SPP1 in pancreatic ductal adenocarcinoma. Genes 13, 1760 (2022).
Yang, J. et al. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol. Life Sci. 77, 305–321 (2020).
Liu, Z.-L., Chen, H.-H., Zheng, L.-L., Sun, L.-P. & Shi, L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct. Target. Ther. 8, 198 (2023).
Curtarello, M. et al. VEGF-targeted therapy stably modulates the glycolytic phenotype of tumor cells. Cancer Res 75, 120–133 (2015).
Lorestani, P. et al. The complex role of macrophages in pancreatic cancer tumor microenvironment: a review on cancer progression and potential therapeutic targets. Discov. Oncol. 15, 369 (2024).
Chen, A. X. et al. Single-cell characterization of macrophages in glioblastoma reveals MARCO as a mesenchymal pro-tumor marker. Genome Med. 13, 88 (2021).
Farhangnia, P., Khorramdelazad, H., Nickho, H. & Delbandi, A.-A. Current and future immunotherapeutic approaches in pancreatic cancer treatment. J. Hematol. Oncol. 17, 40 (2024).
Luo, W., Wen, T. & Qu, X. Tumor immune microenvironment-based therapies in pancreatic ductal adenocarcinoma: time to update the concept. J. Exp. Clin. Cancer Res. 43, 8 (2024).
Joseph, A. M., Al Aiyan, A., Al-Ramadi, B., Singh, S. K. & Kishore, U. Innate and adaptive immune-directed tumour microenvironment in pancreatic ductal adenocarcinoma. Front. Immunol. 15, 1323198 (2024).
Karamitopoulou, E. Tumour microenvironment of pancreatic cancer: immune landscape is dictated by molecular and histopathological features. Br. J. Cancer 121, 5–14 (2019).
Yang, D. et al. Comprehensive analysis of scRNA-Seq and bulk RNA-Seq data reveals dynamic changes in tumor-associated neutrophils in the tumor microenvironment of hepatocellular carcinoma and leads to the establishment of a neutrophil-related prognostic model. Cancer Immunol. Immunother. 72, 4323–4335 (2023).
Wang, Y. et al. Identification of potential immune-related mechanisms related to the development of multiple myeloma. Chin. Med. J. 137, 1603–1613 (2024).
Huo, X. et al. Unravelling the role of immune cells and FN1 in the recurrence and therapeutic process of skull base chordoma. Clin. Transl. Med. 13, e1429 (2023).
Sun, Y. et al. Immunometabolic changes and potential biomarkers in CFS peripheral immune cells revealed by single-cell RNA sequencing. J. Transl. Med. 22, 925 (2024).
Ginhoux, F. & Jung, S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014).
Liu, Z. et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525.e19 (2019).
Mantovani, A. et al. Chemokines in the recruitment and shaping of the leukocyte infiltrate of tumors. Semin Cancer Biol. 14, 155–160 (2004).
Robinson, A., Han, C. Z., Glass, C. K. & Pollard, J. W. Monocyte regulation in homeostasis and malignancy. Trends Immunol. 42, 104–119 (2021).
Qian, B.-Z. et al. FLT1 signaling in metastasis-associated macrophages activates an inflammatory signature that promotes breast cancer metastasis. J. Exp. Med. 212, 1433–1448 (2015).
Dong, Q. et al. MARCO is a potential prognostic and immunotherapy biomarker. Int Immunopharmacol. 116, 109783 (2023).
La Fleur, L. et al. Expression of scavenger receptor MARCO defines a targetable tumor-associated macrophage subset in non-small cell lung cancer. Int J. Cancer 143, 1741–1752 (2018).
Fan, G. et al. The co-location of MARCO+ tumor-associated macrophages and CTSE+ tumor cells determined the poor prognosis in intrahepatic cholangiocarcinoma. Hepatology 82, 25–41 (2025).
Ding, L. et al. Blocking MARCO+ tumor-associated macrophages improves anti-PD-L1 therapy of hepatocellular carcinoma by promoting the activation of STING-IFN type I pathway. Cancer Lett. 582, 216568 (2024).
Sun, H. et al. Association of decreased expression of the macrophage scavenger receptor MARCO with tumor progression and poor prognosis in human hepatocellular carcinoma. J. Gastroenterol. Hepatol. 32, 1107–1114 (2017).
Zhang, B. et al. Development and evaluation of a human CD47/HER2 bispecific antibody for Trastuzumab-resistant breast cancer immunotherapy. Drug Resist. Updates 74, 101068 (2024).
Li, W. et al. Identification of potential resistance mechanisms and therapeutic targets for the relapse of BCMA CAR-T therapy in relapsed/refractory multiple myeloma through single-cell sequencing. Exp. Hematol. Oncol. 12, 44 (2023).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Wang, Z. et al. An immune cell atlas reveals the dynamics of human macrophage specification during prenatal development. Cell 186, 4454–4471.e19 (2023).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020).
Petitprez, F. et al. The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med. 12, 86 (2020).
Xiao, Z., Dai, Z. & Locasale, J. W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 10, 3763 (2019).
Auger, J.-P. et al. Metabolic rewiring promotes anti-inflammatory effects of glucocorticoids. Nature 629, 184–192 (2024).
Li, W. et al. Identification and transcriptome analysis of erythroblastic island macrophages. Blood 134, 480–491 (2019).
Wang, Y. et al. Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. Blood 138, 1615–1627 (2021).
Zhang, Y. et al. Glucocorticoids increase adiposity by stimulating Krüppel-like factor 9 expression in macrophages. Nat. Commun. 15, 1190 (2024).
Acknowledgements
We would like to thank all our authors listed in this manuscript. This work was supported by Natural Science Foundation of China (82400233 (B.Z.), 82270149 (W.L.), 82370144 (Y.S.)); Henan Province Medical Science and Technology Research Project (SBGJ202002087(H.S.), LHGL20240207 (B.Z.)); The Natural Science Foundation of Henan Province (242300421080 (W.L.)); Leading talent project of Henan Province (LJRC2023004, (L.X.)); Postdoctoral Innovative Talent Support Program (GZC20232429(B.Z.)); Henan Provincial Key R&D Program (251111312100 (H.S.)). Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University.
Author information
Authors and Affiliations
Contributions
H.S., M.G., Z.L., and Z.Z. performed experiments, and analyzed the data. B.Z. and S.Z. drafted manuscript. Z.L., Z.Z., T.L., and J.S. analyzed the ScRNA-seq data and bulk-RNA-seq data. H.H., X.L., J.Y., M.S., M.L., Y.A., S.Y., Y.L., Z.H., Y.H., Y.L., C.L., M.L., and M.Y. performed experiments. J.C., Y.S., J.M., M.L., Z.B., and W.L. read and edited the manuscript. L.X., S.Z., B.Z., and G.W. designed and supervised the study and edited the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, H., Gao, M., Liu, Z. et al. Identification and characterization of MARCO-expressing tumor-associated macrophages in pancreatic ductal adenocarcinoma with pan-cancer relevance. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-026-01293-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-026-01293-5