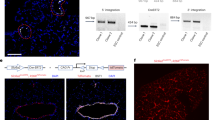
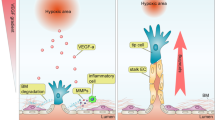
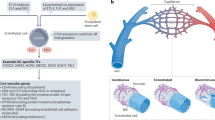
Abstract
The role of the lung’s microcirculation and capillary endothelial cells in normal physiology and the pathobiology of pulmonary diseases is obviously vital. The recent discovery of molecularly distinct aerocytes and general capillary (gCaps) endothelial cells by single-cell transcriptomics (scRNAseq) advanced the field in understanding microcirculatory milieu and cellular communications. However, increasing evidence from different groups indicated the possibility of a more heterogeneous nature of lung capillaries. Therefore, we investigated enriched lung endothelial cells by scRNAseq and identified five novel populations of gCaps with distinct molecular signatures and roles. Our analysis suggests that two major populations of gCaps that express Scn7a(Na+) and Clic4(Cl-) ion transporters form the arterial-to-vein phenotypic transition. We also discovered and named mitotically-active “root” cells (Flot1+ ) on the interface between arterial, Scn7a+ , and Clic4+ endothelium, responsible for the regeneration and repair of the adjacent endothelial populations. Furthermore, the transition of gCaps to a vein requires a venous-capillary endothelium expressing Lingo2. Finally, gCaps disconnected from the zonation represent a high level of Fabp4, other metabolically active genes, and tip-cell markers showing angiogenesis-regulating capacity. The hypoxia-induced models demonstrated that “root” cells exhibit a marked expansion in hypoxia, supporting their role in vascular regeneration and neocapillarization. We also showed a developmental time-course analysis demonstrating an evolution of progenitor (FoxM1+ ) cells, which are progressively replaced by “root” cells during lung maturation, revealing a switch in vascular homeostasis. The discovery of these populations will translate into a better understanding of the involvement of capillary phenotypes and their communications in lung disease pathogenesis.
Similar content being viewed by others
Data availability
All scRNAseq data associated with this manuscript are deposited at Genome Sequence Archive (GSA) with accession ID- PRJCA047802 and submission ID subPRO070142. All other associated data can be found at Dataverse. Fig 1b- https://doi.org/10.7910/DVN/1LSWWO. Fig. 2b- https://doi.org/10.7910/DVN/JNEZCJ, Fig. 4D- https://doi.org/10.7910/DVN/P9PACR, Supplementary data and numerical source data for graphs can be found at https://doi.org/10.7910/DVN/X0VAXH. The raw spatial sequence data (Fig. 5c, d) reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2025) in the National Genomics Data Center (Nucleic Acids Res 2025), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA014847), which are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human/browse/HRA014847.
References
Gillich, A. et al. Capillary cell-type specialization in the alveolus. Nature 586, 785–789 (2020).
Niethamer, T. K. et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife 9, e53072 (2020).
Hogan, B. L. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Koenitzer, J. R., Wu, H., Atkinson, J. J., Brody, S. L. & Humphreys, B. D. Single-Nucleus RNA-Sequencing Profiling of Mouse Lung. Reduced Dissociation Bias and Improved Rare Cell-Type Detection Compared with Single-Cell RNA Sequencing. Am J Respir Cell Mol Biol 63, 739–747 (2020).
D’Armiento, J. Decreased elastin in vessel walls puts the pressure on. The Journal of Clinical Investigation 112, 1308–1310 (2003).
Angel, P. & Karin, M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1072, 129–157 (1991).
Rodor, J. et al. Single-cell RNA sequencing profiling of mouse endothelial cells in response to pulmonary arterial hypertension. Cardiovasc Res 118, 2519–2534 (2022).
Saygin, D. et al. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulm. Circ. 10 https://doi.org/10.1177/2045894020908782 (2020).
The Tabula Sapiens, C. et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896.
Kim, A. J., Alfieri, C. M. & Yutzey, K. E. Endothelial Cell Lineage Analysis Does Not Provide Evidence for EMT in Adult Valve Homeostasis and Disease. Anat Rec (Hoboken) 302, 125–135 (2019).
Pichol-Thievend, C. et al. A blood capillary plexus-derived population of progenitor cells contributes to genesis of the dermal lymphatic vasculature during embryonic development. Development 145 https://doi.org/10.1242/dev.160184 (2018).
Elmasri, H. et al. Endothelial cell-fatty acid binding protein 4 promotes angiogenesis: role of stem cell factor/c-kit pathway. Angiogenesis 15, 457–468 (2012).
del Toro, R. et al. Identification and functional analysis of endothelial tip cell–enriched genes. Blood 116, 4025–4033 (2010).
Ma, J. et al. Activation of JNK/c-Jun is required for the proliferation, survival, and angiogenesis induced by EET in pulmonary artery endothelial cells. J Lipid Res 53, 1093–1105 (2012).
Brown, A. K. & Webb, A. E. Regulation of FOXO Factors in Mammalian Cells. Curr Top Dev Biol 127, 165–192 (2018).
Kim, Y. H. et al. A MST1–FOXO1 cascade establishes endothelial tip cell polarity and facilitates sprouting angiogenesis. Nature Communications 10, 838 (2019).
Otto, D. J., Jordan, C., Dury, B., Dien, C. & Setty, M. Quantifying cell-state densities in single-cell phenotypic landscapes using Mellon. Nat Methods 21, 1185–1195 (2024).
Yuniati, L., Scheijen, B., van der Meer, L. T. & van Leeuwen, F. N. Tumor suppressors BTG1 and BTG2: Beyond growth control. Journal of Cellular Physiology 234, 5379–5389 (2019).
Mahony, R., Ahmed, S., Diskin, C. & Stevenson, N. J. SOCS3 revisited: a broad regulator of disease, now ready for therapeutic use? Cell Mol Life Sci 73, 3323–3336 (2016).
Niethamer, T. K. et al. Atf3 defines a population of pulmonary endothelial cells essential for lung regeneration. Elife 12 https://doi.org/10.7554/eLife.83835 (2023).
Grant, S., Lutz, E. M., McPhaden, A. R. & Wadsworth, R. M. Location and function of VPAC1, VPAC2 and NPR-C receptors in VIP-induced vasodilation of porcine basilar arteries. J Cereb Blood Flow Metab 26, 58–67 (2006).
Hernández-Vásquez, M. N. et al. Cell adhesion controlled by adhesion G protein-coupled receptor GPR124/ADGRA2 is mediated by a protein complex comprising intersectins and Elmo-Dock. J Biol Chem 292, 12178–12191 (2017).
Bochenek, M. L., Dickinson, S., Astin, J. W., Adams, R. H. & Nobes, C. D. Ephrin-B2 regulates endothelial cell morphology and motility independently of Eph-receptor binding. J Cell Sci 123, 1235–1246 (2010).
Chigurupati, S., Kulkarni, T., Thomas, S. & Shah, G. Calcitonin Stimulates Multiple Stages of Angiogenesis by Directly Acting on Endothelial Cells. Cancer Research 65, 8519–8529 (2005).
Young, K. A., Biggins, L. & Sharpe, H. J. Protein tyrosine phosphatases in cell adhesion. Biochem J 478, 1061–1083 (2021).
Madissoon, E. et al. A spatially resolved atlas of the human lung characterizes a gland-associated immune niche. Nat Genet. 55, 66–77 (2023).
Jo, J. H. et al. Novel Gastric Cancer Stem Cell-Related Marker LINGO2 Is Associated with Cancer Cell Phenotype and Patient Outcome. Int J Mol Sci 20 https://doi.org/10.3390/ijms20030555 (2019).
Teng, R. J. et al. Nogo-B receptor modulates angiogenesis response of pulmonary artery endothelial cells through eNOS coupling. Am J Respir Cell Mol Biol 51, 169–177 (2014).
Wälchli, T. et al. Nogo-A is a negative regulator of CNS angiogenesis. Proc Natl Acad Sci USA 110, E1943–E1952 (2013).
Tobia, C. et al. Atypical Chemokine Receptor 3 Generates Guidance Cues for CXCL12-Mediated Endothelial Cell Migration. Front Immunol 10, 1092 (2019).
Pruenster, M. et al. The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nature Immunology 10, 101–108 (2009).
James, J. et al. Heme induces rapid endothelial barrier dysfunction via the MKK3/p38MAPK axis. Blood 136, 749–754 (2020).
Aird, W. C. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med 2, a006429 (2012).
Rodrigues, S. F. & Granger, D. N. Blood cells and endothelial barrier function. Tissue Barriers 3, e978720 (2015).
Komarova, Y. A., Kruse, K., Mehta, D. & Malik, A. B. Protein Interactions at Endothelial Junctions and Signaling Mechanisms Regulating Endothelial Permeability. Circ Res 120, 179–206 (2017).
Chavkin, N. W. & Hirschi, K. K. Single Cell Analysis in Vascular Biology. Front Cardiovasc Med 7, 42 (2020).
Geldhof, V. et al. Single cell atlas identifies lipid-processing and immunomodulatory endothelial cells in healthy and malignant breast. Nature Communications 13, 5511 (2022).
Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris: The Tabula Muris Consortium. Nature 562, 367 (2018).
Belle, N. M. et al. TFF3 interacts with LINGO2 to regulate EGFR activation for protection against colitis and gastrointestinal helminths. Nat Commun 10, 4408 (2019).
Saavedra, P. et al. New insights into circulating FABP4: Interaction with cytokeratin 1 on endothelial cell membranes. Biochim Biophys Acta 1853, 2966–2974 (2015).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nature Communications 8, 14049 (2017).
R: The R Project for Statistical Computing.
Yang, S. et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biology 21, 57 (2020).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Systems 8, 281–291.e289 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Choudhary, S. & Satija, R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biology 23, 27 (2022).
Blanco-Carmona, E. Generating publication ready visualizations for Single Cell transcriptomics using SCpubr. bioRxiv, 2022.2002.2028.482303 https://doi.org/10.1101/2022.02.28.482303.(2022).
Zhao, Q. et al. Single-Cell Transcriptome Analyses Reveal Endothelial Cell Heterogeneity in Tumors and Changes following Antiangiogenic Treatment. Cancer Research 78, 2370–2382 (2018).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biology 20, 59 (2019).
Lange, M. et al. CellRank for directed single-cell fate mapping. Nature Methods 19, 159–170 (2022).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nature Methods 14, 1083–1086 (2017).
vib-singlecell-nf/vsn-pipelines: v0.27.0 v. v0.27.0 (Zenodo, 2021).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLOS ONE 5, e12776 (2010).
Suo, S. et al. Revealing the Critical Regulators of Cell Identity in the Mouse Cell Atlas. Cell Reports 25, 1436–1445.e1433 (2018).
Jones, R. C. et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Saygin, D. et al. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulmonary Circulation 10, 1–15 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nature Communications 12, 1088 (2021).
Sano, T. et al. Image-based crosstalk analysis of cell-cell interactions during sprouting angiogenesis using blood-vessel-on-a-chip. Stem Cell Res Ther 13, 532 (2022).
Otto, D. J., Jordan, C., Dury, B., Dien, C. & Setty, M. Quantifying cell-state densities in single-cell phenotypic landscapes using Mellon. Nature Methods 21, 1185–1195 (2024).
Marsh, S., Salmon, M. & Hoffman, P. samuel-marsh/scCustomize: Version 1.1.1. (Zenodo).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (2016).
wordcloud: Word Clouds v. 2.6 (2018).
Acknowledgements
This work was supported by NIH grants R01HL151447 and R01HL132918 (R.R.), R01HL133085 and R01HL160666 (O.R.), R01HL158596 and R01HL162794 (Z.D.), the AHA postdoctoral fellowship 834220 and NIH K99HL171869 (J.J.) and AHA fellowship 831538 (M.V.), the AHA Transformational award 969574 (OR), the AHA Career Development Award 23CDA1050843 (M.N.). We thank Paulo Pires for the critical reading of the manuscript. We acknowledge the Microscopy Core at Indiana University and the Indiana Center for Biological Microscopy for their support in confocal imaging. We thank the Center for Medical Genomics at Indiana University School of Medicine and the University of Arizona Genetics Core for performing the barcoding and sequencing. We also thank Yukiko T. Matsunaga from the Department of Bioengineering at the University of Tokyo for providing silicone chips used in vessel-on-chip experiments. Additionally, we extend our gratitude to the Pulmonary Hypertension Breakthrough Initiative (grant to PHBI, NHLBI R24 HL123767) for providing tissue OCT blocks for spatial transcriptomics.
Author information
Authors and Affiliations
Contributions
Conception and design: R.R., O.R.; Experimentation, cells isolations, scRNAseq analysis, and interpretation: A.D., O.S.L., N.M., J.J., M.N., M.V., D.B., M.P.R., D.Y., Z.D., H.G., S.K., T.S.; Drafting the manuscript for important intellectual content: R.S.T., O.G., J.J., O.R., R.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
James, J., Dekan, A., Kacar, S. et al. Distinct populations of lung capillary endothelial cells and their functional significance. Commun Biol (2025). https://doi.org/10.1038/s42003-025-09420-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09420-x