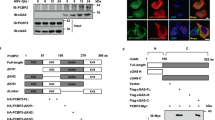
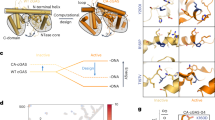
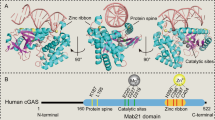
Abstract
The cGAS-STING pathway plays a central role in controlling tumor progression through nucleic acid sensing and type I Interferon production. Here, we identify Poly(rC) Binding Protein 1 as a tumor suppressor that amplifies cGAS-STING signaling in breast cancer. Using patient datasets and a transgenic mouse model with conditional PCBP1 knockout in mammary epithelial cells, we show that PCBP1 expression correlates with improved survival, reduced tumor burden, increased type I Interferon and Interferon Stimulated Gene expression, and elevated cytotoxic T cell infiltration. Mechanistically, PCBP1 binds cytosine-rich single-stranded motifs via its KH domains and increases cGAS affinity to these nucleic acids. Mutation of PCBP1’s conserved GXXG loops impairs nucleic acid binding and cGAS activation. Although cGAS is a double-stranded DNA sensor with no intrinsic sequence specificity, we uncover that the single-stranded nucleic-acid binding protein PCBP1 enhances cGAS sensing by engaging sequence-specific motifs, acting as a nucleic acid co-sensor that impairs tumorigenesis.
Similar content being viewed by others
Data availability
Bulk and single nuclei RNA-sequencing datasets generated during this study are deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE307616 and GSE307750, respectively. Source data are provided with this paper in the supplementary data. The TCGA BRCA dataset is available online at https://www.cancer.gov/tcga. The Tabula Muris dataset is available online at http://tabula-muris.sf.czbiohub.org. Unedited Western blot images are available in the Supplementary Information.
References
Zhang, X. et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430 (2014).
Civril, F. et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013).
Li, X. et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 10.1016 (2013).
Lu, C. et al. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39, 96–108 (2021).
Härtlova, A. et al. DNa damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015).
Kim, J., Kim, H. S. & Chung, J. H. Molecular mechanisms of mitochondrial DNA release and activation of the cGAS-STING pathway. Exp. Mol. Med. 55, 510–519 (2023).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Ablasser, A. et al. CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Hopfner, K. P. & Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS–STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 (2012).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Wang, H. et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. Usa. 114, 1637–1642 (2017).
Li, T. et al. Antitumor activity of cGAMP via stimulation of cGAS-cGAMP-STING-IRF3 mediated innate immune response. Sci. Rep. 12, 19049 (2016).
Vonderhaar, E. P. et al. STING activated tumor-intrinsic type I interferon signaling promotes CXCR3 dependent antitumor immunity in pancreatic cancer. CMGH 12, 41–58 (2021).
Zhang, T. et al. Mesenchymal stromal cells equipped by IFNα empower T cells with potent anti-tumor immunity. Oncogene 41, 1866–1881 (2022).
Mowat, C., Mosley, S. R., Namdar, A., Schiller, D. & Baker, K. Anti-tumor immunity in mismatch repair-deficient colorectal cancers requires type I IFN–driven CCL5 and CXCL10. J. Exp. Med. 218, e20210108 (2021).
Glück, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Hu, J. et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature 616, 806–813 (2023).
Pathare, G. R. et al. Structural mechanism of cGAS inhibition by the nucleosome. Nature 587, 668–672 (2020).
Cao, D., Han, X., Fan, X., Xu, R. M. & Zhang, X. Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res. 30, 1088–1097 (2020).
Kujirai, T. et al. Structural basis for the inhibition of cGAS by nucleosomes. Science 370, 455–458 (2020).
Michalski, S. et al. Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature 587, 678–682 (2020).
Xie, W. et al. Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. USA 116, 11946–11955 (2019).
Andreeva, L. et al. CGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 (2017).
Luecke, S. et al. cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 18, 1707–1715 (2017).
Herzner, A. M. et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 16, 1025–1033 (2015).
Du, Z. et al. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 Å. J. Biol. Chem. 280, 38823–38830 (2005).
Braddock, D. T., Baber, J. L., Levens, D. & Clore, G. M. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 21, 3476–3485 (2002).
Valverde, R., Edwards, L. & Regan, L. Structure and function of KH domains. FEBS J. 275, 2712–2726 (2008).
Dickey, T. H., Altschuler, S. E. & Wuttke, D. S. Single-stranded DNA-binding proteins: multiple domains for multiple functions. Structure 21, 1074–1084 (2013).
Chaudhury, A. et al. TGF-Β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293 (2010).
Grelet, S. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19, 1105–1115 (2017).
Tripathi, V. et al. Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-β. Mol. Cell 64, 549–564 (2016).
Howley, B. V. & Howe, P. H. TGF-beta signaling in cancer: post-transcriptional regulation of EMT via hnRNP E1. Cytokine 118, 19–26 (2019).
Ansa-Addo, E. A. et al. RNA binding protein PCBP1 is an intracellular immune checkpoint for shaping T cell responses in cancer immunity. Sci. Adv. 6 https://doi.org/10.1126/sciadv.aaz3865 (2020).
Shi, H. et al. PCBP1 depletion promotes tumorigenesis through attenuation of p27Kip1 mRNA stability and translation. J. Exp. Clin. Cancer Res. 37, 187 (2018).
Zheng, Y. et al. The RNA-binding protein PCBP1 represses lung adenocarcinoma progression by stabilizing DKK1 mRNA and subsequently downregulating β-catenin. J. Transl. Med. 20, 343 (2022).
Ji, F. J. et al. Expression of both poly r(C) binding protein 1 (PCBP1) and miRNA-3978 is suppressed in peritoneal gastric cancer metastasis. Sci. Rep. 7, 15488 (2017).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020).
Christenson, J. L. et al. MMTV-PyMT and derived Met-1 mouse mammary tumor cells as models for studying the role of the androgen receptor in triple-negative breast cancer progression. Horm. Cancer 8, 69–77 (2017).
Rusinova, I. et al. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41, 1040–1046 (2013).
Ghanem, L. R. et al. The Poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol. Cell. Biol. 36, 304–319 (2016).
Woo, S. R., Corrales, L. & Gajewski, T. F. Innate immune recognition of cancer. Annu. Rev. Immunol. 33, 445–474 (2015).
Fuertes, M. B. et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J. Exp. Med. 208, 2005–2016 (2011).
Diamond, M. S. et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 (2011).
Zitvogel, L., Galluzzi, L., Kepp, O., Smyth, M. J. & Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015).
Liao, C. Y., Lei, C. Q. & Shu, H. B. PCBP1 modulates the innate immune response by facilitating the binding of cGAS to DNA. Cell. Mol. Immunol. 18, 2334–2343 (2021).
Hollingworth, D. et al. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 40, 6873–6886 (2012).
Kranzusch, P. J., Lee, A. S. Y., Berger, J. M. & Doudna, J. A. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 3, 1362–1368 (2013).
Beck, M. A. et al. DNA hypomethylation leads to cGAS-induced autoinflammation in the epidermis. EMBO J. 40, e108234 (2021).
Lai, J. et al. Zebularine elevates STING expression and enhances cGAMP cancer immunotherapy in mice. Mol. Ther. 29, 1758–1771 (2021).
Ng, K.aL. amN. elson et al. Activation of Cgas-sting pathway by hypomethylating agent in TP53 mutated MDS/AML. Blood 144, 2726 (2024).
Michaelis, L. & Menten, M. L. Die Kinetik der Invertinwirkung. Biochem. Z. 49, 333–369 (1913).
Johnson, K. A. & Goody, R. S. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry 50, 8264–8269 (2011).
Grelet, S. et al. TGFβ-induced expression of long noncoding lincRNA Platr18 controls breast cancer axonogenesis. Life Sci. Alliance 5, e202101261 (2022).
Wang, J. et al. VSIG-3 as a ligand of VISTA inhibits human T-cell function. Immunology 156, 74–85 (2019).
Sinha, V. C. et al. Single-cell evaluation reveals shifts in the tumor-immune niches that shape and maintain aggressive lesions in the breast. Nat. Commun. 12, 5024 (2021).
Bai, F. et al. Loss of function of GATA3 induces basal-like mammary tumors. Theranostics 12, 720–733 (2022).
Mohamed, G. A. et al. Lineage plasticity enables low-ER luminal tumors to evolve and gain basal-like traits. Breast Cancer Res. 25, 23 (2023).
Murayama, T. et al. Targeting DHX9 triggers tumor-intrinsic interferon response and replication stress in small cell lung cancer. Cancer Discov. 14, 468–491 (2024).
Ren, X., Liu, Q., Zhou, P., Zhou, T., Wang, D., Mei, Q., Flavell, R. A., Liu, Z., Li, M., Pan, W. & Zhu, S. DHX9 maintains epithelial homeostasis by restraining R-loop-mediated genomic instability in intestinal stem cells. Nat. Commun. 15, 3080 (2024).
Karam, J. A. Q. et al. The RNA-binding protein PCBP1 modulates transcription by recruiting the G-quadruplex-specific helicase DHX9. J. Biol. Chem. 300, 107830 (2024).
Zhang, X. et al. Cyclic GMP-AMP containing mixed Phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013).
Chen, Y. A. et al. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat. Struct. Mol. Biol. 24, 1124–1131 (2017).
Neil, C. et al. The pseudoknot region and poly-(C) tract comprise an essential RNA packaging signal for assembly of foot-and-mouth disease virus. PLoS Pathog. 20, e1012283 (2024).
Martin, L. R., Neal, Z. C., McBride, M. S. & Palmenberg, A. C. Mengovirus and encephalomyocarditis virus poly(c) tract lengths can affect virus growth in murine cell culture. J. Virol. 74, 3074–3081 (2000).
Penza, V., Russell, S. J. & Schulze, A. J. The long-lasting enigma of polycytidine (polyC) tract. PLoS Pathogens 17, e1009739 (2021).
Ryu, M. S., Zhang, D., Protchenko, O., Shakoury-Elizeh, M. & Philpott, C. C. PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J. Clin. Invest. 127, 1786–1797 (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Germain, P. L., Lun, A., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Research 10, 979 (2021).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods 16, 1289–1296 (2019).
Alquicira-Hernandez, J., Sathe, A., Ji, H. P., Nguyen, Q. & Powell, J. E. ScPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 20, 264 (2019).
Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Squair, J. W. et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 12, 5692 (2021).
Acknowledgements
We would like to thank all members of Dr. Philip Howe’s laboratory, Dr. Shikhar Mehrotra, and Paramita Chakraborty for their technical help and helpful feedback. We also thank members of the Hollings Cancer Centre Translational Science Shared Resource, the Bioinformatics Core and the Flow Cytometry & Cell Sorting Shared Resource. This work was supported by the National Institutes of Health, National Cancer Institute grant [CA154663] to P.H.H. The work was also supported in part by the Translational Science Shared Resource, Bioinformatics Core and Flow Cytometry & Cell Sorting Shared Resource, Hollings Cancer Centre, Medical University of South Carolina [P30 CA138313]. Funding for open access charge: National Institutes of Health.
Author information
Authors and Affiliations
Contributions
C.F. conceived the study, designed the methodology, acquired all data, and wrote the manuscript. J.A.Q.K. assisted with EMSAs, and B.M. contributed additional methodological support. P.C. provided expertise in methodology and acquisition of flow cytometry data, with resource support from S.M. Library preparation for bulk and single-nuclei RNA sequencing was performed by S.V. and Romeo Martin. Bioinformatic analysis of bulk RNA sequencing was carried out by C.F., and single-nuclei RNA sequencing analysis by B.G. A.D. analysed the CD8/CD3 multiplex IHC. Additional supervision was provided by B.V.H. and P.H.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Francisco Venegas Solis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Isabela Pedroza-Pacheco and Johannes Stortz. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fréreux, C., Karam, J.A.Q., Howley, B.V. et al. PCBP1 binding to single-stranded poly-cytosine motifs enhances cGAS sensing and impairs breast cancer development. Commun Biol (2026). https://doi.org/10.1038/s42003-025-09456-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09456-z