Abstract
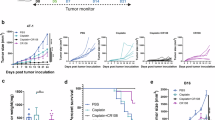
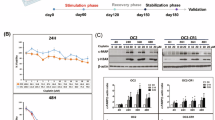
Liver cancer treatment with cisplatin is often hindered by drug resistance. This study aimed to identify key genes associated with cisplatin resistance in liver cancer and develop targeted inhibitors. Using genome-wide CRISPR-Cas9 screening, ATOX1 was identified as a critical gene for cisplatin resistance. ATOX1 was highly expressed in liver cancer tissues and associated with poor prognosis. Knockdown of ATOX1 in liver cancer cells enhanced cisplatin sensitivity in vitro and in vivo. Molecular dynamics simulation and virtual screening identified compound 8 as a potent ATOX1 inhibitor with high affinity (Kd = 12.5 μM) and exhibited synergistic effects with cisplatin on liver cancer cell growth. Mechanistically, compound 8 inhibits the activity of ATOX1, leading to intracellular copper accumulation. The elevated copper levels subsequently promote increased DNA methylation at the NOTCH1 promoter, resulting in suppression of the NOTCH1/HES1 signaling pathway and enhancing the sensitivity of liver cancer cells to cisplatin. In conclusion, ATOX1 is crucial for cisplatin resistance in liver cancer and linked to poor prognosis. Targeting ATOX1 with compound 8 may be a novel therapeutic strategy for overcoming cisplatin resistance.
Similar content being viewed by others
Data availability
The raw FASTQ files from the CRISPR/Cas9 library screen and RNA-seq have been deposited in the CNCB database (https://www.cncb.ac.cn/) under accession number PRJCA056149. All other raw data are provided in the Supplementary Data and are available from the corresponding author upon reasonable request.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Rumgay, H. et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 77, 1598–1606 (2022).
Oh, J. H. & Jun, D. W. The latest global burden of liver cancer: a past and present threat. Clin. Mol. Hepatol. 29, 355–357 (2023).
Forner, A., Reig, M. & Bruix, J. Hepatocellular carcinoma. Lancet 391, 1301–1314 (2018).
Feng, F. & Zhao, Y. Hepatocellular carcinoma: prevention, diagnosis, and treatment. Med. Princ. Pract. 33, 414–423 (2024).
Dasari, S. & Tchounwou, P. B. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 740, 364–378 (2014).
Galluzzi, L. et al. Molecular mechanisms of cisplatin resistance. Oncogene 31, 1869–1883 (2012).
Siddik, Z. H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279 (2003).
Petruzzelli, R. & Polishchuk, R. S. Activity and trafficking of copper-transporting ATPases in tumor development and defense against platinum-based drugs. Cells 8, 108 (2019).
Li, Y. Q. et al. Copper efflux transporters ATP7A and ATP7B: novel biomarkers for platinum drug resistance and targets for therapy. IUBMB Life 70, 183–191 (2018).
Boal, A. K. & Rosenzweig, A. C. Crystal structures of cisplatin bound to a human copper chaperone. J. Am. Chem. Soc. 131, 14196–14197 (2009).
Itoh, S. et al. Novel role of antioxidant-1 (Atox1) as a copper-dependent transcription factor involved in cell proliferation. J. Biol. Chem. 283, 9157–9167 (2008).
Blockhuys, S., Zhang, X. & Wittung-Stafshede, P. Single-cell tracking demonstrates copper chaperone Atox1 to be required for breast cancer cell migration. Proc. Natl. Acad. Sci. USA 117, 2014–2019 (2020).
Zhao, X. et al. Expression of cuproptosis-related genes in hepatocellular carcinoma and their relationships with prognosis. Front. Oncol. 12, 992468 (2022).
Boal, A. K. & Rosenzweig, A. C. Structural biology of copper trafficking. Chem. Rev. 109, 4760–4779 (2009).
Palm-Espling, M. E. et al. Determinants for simultaneous binding of copper and platinum to human chaperone Atox1: hitchhiking not hijacking. PLoS ONE 8, e70473 (2013).
Arnesano, F. et al. Characterization of the binding interface between the copper chaperone Atx1 and the first cytosolic domain of Ccc2 ATPase. J. Biol. Chem. 276, 41365–41376 (2001).
Palm, M. E. et al. Cisplatin binds human copper chaperone Atox1 and promotes unfolding in vitro. Proc. Natl. Acad. Sci. USA 108, 6951–6956 (2011).
Safaei, R. et al. Role of copper transporters in the development of resistance to platinum drugs. J. Inorg. Biochem. 98, 1607–1613 (2004).
Xie, J., Yang, Y., Gao, Y. & He, J. Cuproptosis: mechanisms and links with cancers. Mol. Cancer 22, 46 (2023).
Kong, R. & Sun, G. Targeting copper metabolism: a promising strategy for cancer treatment. Front. Pharmacol. 14, 1203447 (2023).
Huang, X., Lian, M. & Li, C. Copper homeostasis and cuproptosis in gynecological cancers. Front. Cell Dev. Biol. 12, 1459183 (2024).
Li, Z. H. et al. Role of copper transporter ATP7A in platinum resistance of esophageal squamous cell carcinoma. J. Cancer 7, 2085–2092 (2016).
Guan, D. et al. Copper in cancer: from pathogenesis to therapy. Biomed. Pharmacother. 163, 114791 (2023).
Li, J. L. & Harris, A. L. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer Cell 8, 1–3 (2005).
Maharati, A. & Moghbeli, M. Forkhead box proteins as critical regulators of cisplatin response in tumor cells. Eur. J. Pharmacol. 956, 175937 (2023).
Inkol, J. M., Poon, A. C. & Mutsaers, A. J. Inhibition of copper chaperones sensitizes human and canine osteosarcoma cells to carboplatin chemotherapy. Vet. Comp. Oncol. 18, 559–569 (2020).
Wang, J. et al. Inhibition of human copper trafficking by a small molecule attenuates cancer cell proliferation. Nat. Chem. 7, 968–979 (2015).
Fu, L. et al. Current research on the Notch pathway in hepatocellular carcinoma. Eur. J. Med. Res. 30, 402 (2025).
Sosa Iglesias, V. et al. Drug resistance in non-small cell lung cancer: a potential for NOTCH targeting? Front. Oncol 8, 267 (2018).
Elkin, E. R. et al. Metals exposures and DNA methylation: current evidence and future directions. Curr. Environ. Health Rep. 9, 673–696 (2022).
Santos, D. et al. Microplastics- and copper-induced changes in neurogenesis and DNA methyltransferases in early life stages of zebrafish. Chem. Biol. Interact. 363, 110021 (2022).
Makovec, T. Cisplatin and beyond: molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiol. Oncol. 53, 148–158 (2019).
Howell, S. B. et al. Copper transporters and the cellular pharmacology of platinum-containing cancer drugs. Mol. Pharmacol. 77, 887–894 (2010).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Galluzzi, L. et al. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 5, e1257 (2014).
Easwaran, H., Tsai, H. C. & Baylin, S. B. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 54, 716–727 (2014).
Galluzzi, L. et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 8, e000337 (2020).
Vasan, N., Baselga, J. & Hyman, D. M. A view on drug resistance in cancer. Nature 575, 299–309 (2019).
Rottenberg, S. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. USA 105, 17079–17084 (2008).
Dai, Y. et al. Correction: involvement of Bcl-2 family proteins in AKT-regulated cell survival in cisplatin-resistant epithelial ovarian cancer. Oncotarget 11, 488–489 (2020).
Denoyer, D. et al. Targeting copper in cancer therapy: “copper that cancer. Metallomics 7, 1459–1476 (2015).
Yuan, X. et al. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer Lett. 369, 20–27 (2015).
Wang, Z. et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim. Biophys. Acta 1806, 258–267 (2010).
Liu, J. et al. NOTCH1 regulates DNA damage response and sorafenib resistance by activating ATM in hepatocellular carcinoma. Am. J. Transl. Res. 16, 7317–7329 (2024).
Chen, Z. X. et al. Hypoxia-induced DTL promotes proliferation, metastasis, and sorafenib resistance of hepatocellular carcinoma via ubiquitin-mediated degradation of SLTM and activation of Notch signaling. Cell Death Dis. 15, 734 (2024).
Adamowicz, M., Vermezovic, J. & d’Adda di Fagagna, F. NOTCH1 inhibits activation of ATM by impairing formation of an ATM–FOXO3a–KAT5/Tip60 complex. Cell Rep. 16, 2068–2076 (2016).
Tong, X. et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis. J. Hematol. Oncol. 15, 174 (2022).
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (82560795, 82260535), Guizhou Provincial Science and Technology Projects General (ZK[2026]341). We would like to thank the Figdraw platform (https://www.figdraw.com/#/) for providing the original materials and authorization approval for the creation of the schematic diagrams for Figs. 1A and 5G. The authorization approval numbers are TWWYA9335b and UWPIIde6be, respectively.
Author information
Authors and Affiliations
Contributions
Dan Ma, Shi Zuo, Lei Tang, and Zhirui Zeng designed or supported the research and revised the manuscript. Chujiao Hu wrote the paper. Chujiao Hu, Huading Tai, Renguang Zhu, and Zhirui Zeng performed the research and analyzed the data. Chujiao Hu, Huading Tai, Renguang Zhu, Zhirui Zeng, Zhengyu Shu, and Guanghao Guo participated in animal experiments. All the authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All authors consent to the publication of this manuscript. Individual data included in this study are anonymized.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Joanna Timmins and Mengtan Xing. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, C., Tai, H., Zhu, R. et al. CRISPR-Cas9 screening identifies ATOX1-driven cisplatin resistance mechanisms in liver cancer and evaluates targeted inhibitor efficacy. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09722-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09722-8