Abstract
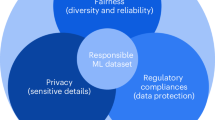
Artificial intelligence (AI) tools are increasingly being used to help make consequential decisions about individuals. While AI models may be accurate on average, they can simultaneously be highly uncertain about outcomes associated with specific individuals or groups of individuals. For high-stakes applications (such as healthcare and medicine, defence and security, banking and finance), AI decision-support systems must be able to make personalized assessments of uncertainty in a rigorous manner. However, the statistical frameworks needed to do so are currently incomplete. Here, we outline current approaches to personalized uncertainty quantification (PUQ) and define a set of grand challenges associated with the development and use of PUQ in a range of areas, including multimodal AI, explainable AI, generative AI and AI fairness.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Allen, B. The promise of explainable AI in digital health for precision medicine: a systematic review. J. Personalized Med. 14, 277 (2024).
Battaglini, M. & Rasmussen, S. Transparency, automated decision-making processes and personal profiling. J. Data Prot. Priv. 2, 331–349 (2019).
Christensen, J. in Demystifying AI for the Enterprise 149–192 (Productivity, 2021).
Hüllermeier, E. & Waegeman, W. Aleatoric and epistemic uncertainty in machine learning: an introduction to concepts and methods. Mach. Learn. 110, 457–506 (2021).
Der Kiureghian, A. & Ditlevsen, O. Aleatory or epistemic? Does it matter? Struct. Saf. 31, 105–112 (2009).
Volodina, V. & Challenor, P. The importance of uncertainty quantification in model reproducibility. Philos. Trans. R. Soc. A 379 (2021).
Walther, B. A. & Moore, J. L. The concepts of bias, precision and accuracy, and their use in testing the performance of species richness estimators, with a literature review of estimator performance. Ecography 28, 815–829 (2005).
Krishnan, R. & Tickoo, O. Improving model calibration with accuracy versus uncertainty optimization. Adv. Neural Inf. Process. Syst. 33, 18237–18248 (2020).
Rogers, W. A. & Walker, M. J. Fragility, uncertainty, and healthcare. Theor. Med. Bioeth. 37, 71–83 (2016).
Podkopaev, A. & Ramdas, A. Distribution-free uncertainty quantification for classification under label shift. Proc. Mach. Learn. Res. 161, 844–853 (2021).
Thuy, A. & Benoit, D. F. Explainability through uncertainty: trustworthy decision-making with neural networks. Eur. J. Oper. Res. 317, 330–340 (2024).
Kumar, S. & Srivastava, A. Bootstrap prediction intervals in non-parametric regression with applications to anomaly detection. In Proc. 18th ACM SIGKDD Conference on Knowledge Discovery and Data Mining (2012).
Gal, Y. & Ghahramani, Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. Proc. Mach. Learn. Res. 48, 1050–1059 (2016).
Milanés-Hermosilla, D. et al. Monte Carlo dropout for uncertainty estimation and motor imagery classification. Sensors 21, 7241 (2021).
Kotelevskii, N., Horváth, S., Nandakumar, K., Takáč, M. & Panov, M. Dirichlet-based uncertainty quantification for personalized federated learning with improved posterior networks. In Proc. Thirty-Third International Joint Conference on Artificial Intelligence (IJCAI-24) 7127–7135 (2023).
Wang, X. & Kadıoğlu, S. Modeling uncertainty to improve personalized recommendations via bayesian deep learning. Int. J. Data Sci. Anal. 16, 191–201 (2023).
Vovk, V., Gammerman, A. & Shafer, G. Algorithmic Learning in a Random World Vol. 29 (Springer, 2005).
Angelopoulos, A. N. & Bates, S. A gentle introduction to conformal prediction and distribution-free uncertainty quantification. Foundations and Trends in Machine Learning 16, 494–591 (2023).
Papadopoulos, H., Proedrou, K., Vovk, V. & Gammerman, A. Inductive confidence machines for regression. In Machine Learning: ECML 2002 (eds Elomaa, T. et al.) 345–356 (Lecture Notes in Computer Science Vol. 2430, Springer, 2002).
Barber, R. F., Candes, E. J., Ramdas, A. & Tibshirani, R. J. Conformal prediction beyond exchangeability. Ann. Stat. 51, 816–845 (2023).
Gibbs, I. & Candes, E. Adaptive conformal inference under distribution shift. Adv. Neural Inf. Process. Syst. 34, 1660–1672 (2021).
Gibbs, I. & Candes, E. Conformal inference for online prediction with arbitrary distribution shifts. J. Mach. Learn. Res. 25.162, 1–36 (2024).
Angelopoulos, A. N., Barber, R. F. & Bates, S. Online conformal prediction with decaying step sizes. Proc. Mach. Learn. Res. 235, 1616–1630 (2024).
Zaffran, M., Féron, O., Goude, Y., Josse, J. & Dieuleveut, A. Adaptive conformal predictions for time series. Proc. Mach. Learn. Res. 162, 25834–25866 (2022).
Lei, J. & Wasserman, L. Distribution-free prediction bands for non-parametric regression. J. R. Stat. Soc. B 76, 71–96 (2014).
Löfström, T., Boström, H., Linusson, H. & Johansson, U. Bias reduction through conditional conformal prediction. Intell. Data Anal. 19, 1355–1375 (2015).
Foygel Barber, R., Candes, E. J., Ramdas, A. & Tibshirani, R. J. The limits of distribution-free conditional predictive inference. Inf. Inference 10, 455–482 (2021).
Romano, Y., Patterson, E. & Candes, E. Conformalized quantile regression. Adv. Neural Inf. Process. Syst. 32 (2019).
Romano, Y., Sesia, M. & Candes, E. Classification with valid and adaptive coverage. Adv. Neural Inf. Process. Syst. 33, 3581–3591 (2020).
Guan, L. Localized conformal prediction: a generalized inference framework for conformal prediction. Biometrika 110, 33–50 (2023).
Ding, T., Angelopoulos, A., Bates, S., Jordan, M. & Tibshirani, R. J. Class-conditional conformal prediction with many classes. Adv. Neural Inf. Process. Syst. 36 (2024).
Romano, Y., Barber, R. F., Sabatti, C. & Candès, E. With malice toward none: assessing uncertainty via equalized coverage. Harv. Data Sci. Rev. 2, 4 (2020).
Sadinle, M., Lei, J. & Wasserman, L. Least ambiguous set-valued classifiers with bounded error levels. J. Am. Stat. Assoc. 114, 223–234 (2019).
Gibbs, I., Cherian, J. J. & Candès, E. J. Conformal prediction with conditional guarantees. Journal of the Royal Statistical Society Series B: Statistical Methodology, 2025.
Ye, D. et al. Uncertainty quantification patterns for multiscale models. Philos. Trans. R. Soc. A 379, 20200072 (2021).
Akbari, A. & Jafari, R. Personalizing activity recognition models through quantifying different types of uncertainty using wearable sensors. IEEE Trans. Biomed. Eng. 67, 2530–2541 (2020).
Fontana, M., Zeni, G. & Vantini, S. Conformal prediction: a unified review of theory and new challenges. Preprint at https://arxiv.org/abs/2005.07972 (2021).
Abdar, M. et al. A review of uncertainty quantification in deep learning: techniques, applications and challenges. Inf. Fusion 76, 243–297 (2021).
Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018).
Ji, Q. et al. Multimodal omics approaches to aging and age-related diseases. Phenomics 4, 56–71 (2024).
Canali, S., Schiaffonati, V. & Aliverti, A. Challenges and recommendations for wearable devices in digital health: data quality, interoperability, health equity, fairness. PLoS Digit. Health 1, e0000104 (2022).
Futoma, J. et al. As good as it gets? A new approach to estimating possible prediction performance. PLoS ONE 19, e0296904 (2024).
Boehm, K. M., Khosravi, P., Vanguri, R., Gao, J. & Shah, S. P. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer 22, 114–126 (2022).
Baltrušaitis, T., Ahuja, C. & Morency, L.-P. Multimodal machine learning: a survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 41, 423–443 (2018).
Rahate, A., Walambe, R., Ramanna, S. & Kotecha, K. Multimodal co-learning: challenges, applications with datasets, recent advances and future directions. Inf. Fusion 81, 203–239 (2022).
Longo, L. et al. Explainable Artificial Intelligence (XAI) 2.0: a manifesto of open challenges and interdisciplinary research directions. Inf. Fusion 106, 102301 (2024).
Ayci, G., Sensoy, M., Özgür, A. & Yolum, P. Uncertainty-aware personal assistant for making personalized privacy decisions. ACM Trans. Internet Technol. 23, 13 (2023).
Tjoa, E. & Guan, C. A survey on Explainable Artificial Intelligence (XAI): toward medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 32, 4793–4813 (2020).
Qian, W. et al. Towards modeling uncertainties of self-explaining neural networks via conformal prediction. In Proc. AAAI Conference on Artificial Intelligence Vol. 38 14651–14659 (2024).
Almaslukh, B. A reliable breast cancer diagnosis approach using an optimized deep learning and conformal prediction. Biomed. Signal Process. Control 98, 106743 (2024).
Quinonero-Candela, J. et al. Dataset Shift in Machine Learning (MIT Press, 2009).
Mougan, C. & Nielsen, D. Monitoring model deterioration with explainable uncertainty estimation via non-parametric bootstrap. In Proc. AAAI Conference on Artificial Intelligence Vol. 37:12 15037–15045 (2023).
Gama, J., Žliobaitė, I., Bifet, A., Pechenizkiy, M. & Bouchachia, A. A survey on concept drift adaptation. ACM Comput. Surv. 46, 44 (2014).
Sebastiao, R. & Gama, J. On evaluating stream learning algorithms. Mach. Learn. 90, 317–346 (2013).
Sebastiao, R. & Gama, J. A study on change detection methods. In Progress in Artificial Intelligence, 14th Portuguese Conference on Artificial Intelligence, EPIA 12–15 (2009).
Bayram, F., Ahmed, B. & Kassler, A. From concept drift to model degradation: an overview on performance-aware drift detectors. Knowl.-Based Syst. 245, 108632 (2022).
Garg, S., Balakrishnan, S., Lipton, Z., Neyshabur, B. & Sedghi, H. Leveraging unlabeled data to predict out-of-distribution performance. In NeurIPS 2021 Workshop on Distribution Shifts: Connecting Methods and Applications (2021).
Barber, R., Candes, E., Ramdas, A. & Tibshirani, R. Predictive inference with the jackknife+. Ann. Stat. 49, 486–507 (2021).
Tibshirani, R. J., Foygel Barber, R., Candes, E. & Ramdas, A. Conformal prediction under covariate shift. Adv. Neural Inf. Process. Syst. 32 (2019).
de Barros, R. & de Carvalho Santos, S. An overview and comprehensive comparison of ensembles for concept drift. Inf. Fusion 52, 213–244 (2019).
Barros, R. & Santos, S. A large-scale comparison of concept drift detectors. Inf. Sci. 451, 348–370 (2018).
Ovadia, Y. et al. Can you trust your model’s uncertainty? Evaluating predictive uncertainty under dataset shift. Proceedings of NeurIPS (2019).
Lobo, J., Del Ser, J., Bifet, A. & Kasabov, N. Spiking neural networks and online learning: an overview and perspectives. Neural Netw. 121, 88–100 (2020).
Cao, Y. et al. Knowledge-preserving incremental social event detection via heterogeneous GNNs. In Proc. Web Conference 2021 3383–3395 (2021).
Mitra, R. et al. Learning from data with structured missingness. Nat. Mach. Intell. 5, 13–23 (2023).
Jackson, J., Mitra, R., Hagenbuch, N., McGough, S. & Harbron, C. A complete characterisation of structured missingness. Preprint at https://arxiv.org/abs/2307.02650 (2023).
Bianconi, G. Multilayer Networks (Oxford Univ. Press, 2018).
Bianconi, G. Higher-Order Networks (Cambridge Univ. Press, 2021).
Baptista, A., Sánchez-García, R. J., Baudot, A. & Bianconi, G. Zoo guide to network embedding. Journal of Physics: Complexity. 4 (2023).
Gutknecht, A. J., Wibral, M. & Makkeh, A. Bits and pieces: understanding information decomposition from part–whole relationships and formal logic. Proc. R. Soc. A 477, 20210110 (2021).
Kiang, M. V. et al. Sociodemographic characteristics of missing data in digital phenotyping. Sci. Rep. 11, 14447 (2021).
Tsiampalis, T. & Panagiotakos, D. B. Missing-data analysis: socio-demographic, clinical and lifestyle determinants of low response rate on self-reported psychological and nutrition related multi-item instruments in the context of the ATTICA epidemiological study. BMC Med. Res. Methodol. 20, 148 (2020).
Angwin, J., Larson, J., Mattu, S. & Kirchner, L. Machine bias. ProPublica https://www.propublica.org/article/machine-bias-risk-assessments-in-criminal-sentencing (2016).
Vyas, D. A., Eisenstein, L. G. & Jones, D. S. Hidden in plain sight—reconsidering the use of race correction in clinical algorithms. N. Engl. J. Med. 382, 2465–2474 (2020).
Ibrahim, H., Liu, X., Zariffa, N., Morris, A. D. & Denniston, A. K. Health data poverty: an assailable barrier to equitable digital health care. Lancet Digit. Health 2, E666–E676 (2020).
Fatumo, S. et al. A roadmap to increase diversity in genomic studies. Nat. Med. 27, 24–29 (2021).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 1565–1576 (2019).
Buolamwini, J. & Gebru, T. Gender shades: intersectional accuracy disparities in commercial gender classification. In Proc. 1st Conference on Fairness, Accountability and Transparency 77–91 (2018).
Seyyed-Kalantari, L., Zhang, H., McDermott, M. B. A., Chen, I. Y. & Ghassemi, M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat. Med. 27, 111–120 (2021).
Chen, I. Y., Johansson, F. D. & Sontag, D. Why is my classifier discriminatory? In Proc. NIPS 3543–3554 (2018).
Matovu, E. et al. Enabling the genomic revolution in Africa. Science 344, 1260792 (2014).
Suriyakumar, V. & Narayanan, A. Proc. Mach. Learn. Res. 202, 7395–7405 (2023).
Régis, C., Denis, J.-L., Axente, M. L. & Kishimoto, A. Human-Centered AI: a Multidisciplinary Perspective for Policy-Makers, Auditors, and Users (Chapman & Hall, 2024).
Combs, K., Moyer, A. & Bihl, T. J. Uncertainty in visual generative AI. Algorithms 17, 136 (2024).
Farquhar, S., Kossen, J., Kuhn, L. & Gal, Y. Detecting hallucinations in large language models using semantic entropy. Nature 630, 625–630 (2024).
Kanwal, N. et al. Are you sure it’s an artifact? Artifact detection and uncertainty quantification in histological images. Comput. Med. Imaging Graph. 112, 102321 (2024).
Zhou, E. & Lee, D. Generative artificial intelligence, human creativity, and art. PNAS nexus 3, pgae052 (2024).
Grant, N. Google chatbot’s A.I. images put people of color in Nazi-era uniforms. New York Times https://www.nytimes.com/2024/02/22/technology/google-gemini-german-uniforms.html (2024).
Subramanian, H. V., Canfield, C., Shank, D. B. & Kinnison, M. Combining uncertainty information with AI recommendations supports calibration with domain knowledge. J. Risk Res. 26, 1137–1152 (2023).
Harish, V., Morgado, F., Stern, A. D. & Das, S. Artificial intelligence and clinical decision making: the new nature of medical uncertainty. Acad. Med. 96, 31–36 (2021).
DeFrank, J. & Luiz, A. AI-based personalized treatment recommendation for cancer patients. J. Carcinogenesis 21, 57–63 (2022).
European Union. Regulation (EU) 2024/1689. Official J. Eur. Union (2024).
Acknowledgements
This work is supported by the Turing–Roche Strategic Partnership. C.R.S.B. was supported by the CRUK City of London Centre Award (CTRQQR-2021\100004). A.F.F. acknowledges support from the Royal Academy of Engineering under the RAEng Chair in Emerging Technologies (INSILEX CiET1919\/19), ERC Advanced Grant – UKRI Frontier Research Guarantee (INSILICO EP\/Y030494/1). A.F.F. also acknowledges the National Institute for Health and Care Research (NIHR) Manchester Biomedical Research Centre (BRC) (NIHR203308). T.C. is supported by a principal research fewllowship from the UCL Biomedical Research Centre (BRC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakraborti, T., Banerji, C.R.S., Marandon, A. et al. Personalized uncertainty quantification in artificial intelligence. Nat Mach Intell 7, 522–530 (2025). https://doi.org/10.1038/s42256-025-01024-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01024-8
This article is cited by
-
Multimodal out-of-distribution individual uncertainty quantification enhances binding affinity prediction for polypharmacology
Nature Machine Intelligence (2025)
-
Deciphering authenticity in the age of AI: how AI-generated disinformation images and AI detection tools influence judgements of authenticity
AI & SOCIETY (2025)
-
Conformal uncertainty quantification to evaluate predictive fairness of foundation AI model for skin lesion classes across patient demographics
Health Information Science and Systems (2025)
-
Trust and Uncertainties: Characterizing Trustworthy AI Systems Within a Multidimensional Theory of Trust
Topoi (2025)
-
Artificial intelligence for TNM staging in NSCLC: a critical appraisal of segmentation utility in [1⁸F]FDG PET/CT
European Journal of Nuclear Medicine and Molecular Imaging (2025)