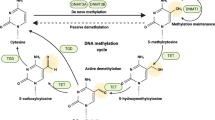
Abstract
Epigenetic dysregulation is increasingly appreciated as a hallmark of cancer, including disease initiation, maintenance and therapy resistance. As a result, there have been advances in the development and evaluation of epigenetic therapies for cancer, revealing substantial promise but also challenges. Three epigenetic inhibitor classes are approved in the USA, and many more are currently undergoing clinical investigation. In this Review, we discuss recent developments for each epigenetic drug class and their implications for therapy, as well as highlight new insights into the role of epigenetics in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Mashtalir, N. et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183, 802–817 (2020).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Lai, W. K. M. & Pugh, B. F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017).
Mittal, P. & Roberts, C. W. M. The SWI/SNF complex in cancer—biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435–448 (2020).
Cramer, P. Organization and regulation of gene transcription. Nature 573, 45–54 (2019).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Conery, A. R., Rocnik, J. L. & Trojer, P. Small molecule targeting of chromatin writers in cancer. Nat. Chem. Biol. 18, 124–133 (2022).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
Dharia, N. V. et al. A first-generation pediatric cancer dependency map. Nat. Genet. 53, 529–538 (2021).
Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 (2018).
Baylin, S. B. & Jones, P. A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a019505 (2016).
Whittaker, S. J. et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 28, 4485–4491 (2010).
O’Connor, O. A. et al. Belinostat in patients with relapsed or refractory peripheral T-cell lymphoma: results of the pivotal phase II BELIEF (CLN-19) study. J. Clin. Oncol. 33, 2492–2499 (2015).
San-Miguel, J. F. et al. Overall survival of patients with relapsed multiple myeloma treated with panobinostat or placebo plus bortezomib and dexamethasone (the PANORAMA 1 trial): a randomised, placebo-controlled, phase 3 trial. Lancet Haematol. 3, e506–e515 (2016).
Eckschlager, T., Plch, J., Stiborova, M. & Hrabeta, J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. https://doi.org/10.3390/ijms18071414 (2017).
Lechner, S. et al. Target deconvolution of HDAC pharmacopoeia reveals MBLAC2 as common off-target. Nat. Chem. Biol. 18, 812–820 (2022).
Najm, F. J. et al. Chromatin complex dependencies reveal targeting opportunities in leukemia. Nat. Commun. 14, 448 (2023).
Zhang, Y. et al. Collateral lethality between HDAC1 and HDAC2 exploits cancer-specific NuRD complex vulnerabilities. Nat. Struct. Mol. Biol. 30, 1160–1171 (2023).
Chang, L., Ruiz, P., Ito, T. & Sellers, W. R. Targeting pan-essential genes in cancer: challenges and opportunities. Cancer Cell 39, 466–479 (2021).
Yang, Z. et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545 (2005).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Puissant, A. et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 (2013).
Hensel, T. et al. Targeting the EWS–ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget 7, 1451–1463 (2016).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Doroshow, D. B., Eder, J. P. & LoRusso, P. M. BET inhibitors: a novel epigenetic approach. Ann. Oncol. 28, 1776–1787 (2017).
Piha-Paul, S. A. et al. Phase 1 study of molibresib (GSK525762), a bromodomain and extra-terminal domain protein inhibitor, in NUT carcinoma and other solid tumors. JNCI Cancer Spectr. 4, pkz093 (2020).
Stathis, A. et al. Clinical response of carcinomas harboring the BRD4–NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 6, 492–500 (2016).
Shorstova, T., Foulkes, W. D. & Witcher, M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 124, 1478–1490 (2021).
Sun, Y. et al. Safety and efficacy of bromodomain and extra-terminal inhibitors for the treatment of hematological malignancies and solid tumors: a systematic study of clinical trials. Front. Pharmacol. 11, 621093 (2020).
Garraway, L. A. & Lander, E. S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Morschhauser, F. et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 21, 1433–1442 (2020).
Ogiwara, H. et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 6, 430–445 (2016).
Scheller, M. et al. Hotspot DNMT3A mutations in clonal hematopoiesis and acute myeloid leukemia sensitize cells to azacytidine via viral mimicry response. Nat. Cancer 2, 527–544 (2021).
Bejar, R. et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 124, 2705–2712 (2014).
Gounder, M. et al. Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: an international, open-label, phase 2 basket study. Lancet Oncol. 21, 1423–1432 (2020).
Czermin, B. et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 (2002).
Müller, J. et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 (2002).
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
McKinney, M. et al. The genetic basis of hepatosplenic T-cell lymphoma. Cancer Discov. 7, 369–379 (2017).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Pikman, Y. et al. Targeting EZH2 for the treatment of hepatosplenic T-cell lymphoma. Blood Adv. 4, 1265–1269 (2020).
Tirode, F. et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4, 1342–1353 (2014).
Chen, L. et al. CRISPR–Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J. Clin. Invest. 128, 446–462 (2018).
Duan, R., Du, W. & Guo, W. EZH2: a novel target for cancer treatment. J. Hematol. Oncol. 13, 104 (2020).
Qi, W. et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl Acad. Sci. USA 109, 21360–21365 (2012).
Richter, G. H. S. et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl Acad. Sci. USA 106, 5324–5329 (2009).
Wilson, B. G. et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010).
Kadoch, C. et al. Dynamics of BAF–Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 49, 213–222 (2017).
Bitler, B. G. et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 21, 231–238 (2015).
Knutson, S. K. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. USA 110, 7922–7927 (2013).
Ntziachristos, P. et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 18, 298–301 (2012).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726 (2010).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42, 665–667 (2010).
Lee, W. et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 46, 1227–1232 (2014).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Chi, S. et al. Abstract A175: phase 1 study of the EZH2 inhibitor, tazemetostat, in children with relapsed or refractory INI1-negative tumors including rhabdoid tumors, epithelioid sarcoma, chordoma, and synovial sarcoma. Mol. Cancer Ther. 17, A175 (2018).
Honma, D. et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 108, 2069–2078 (2017).
Ribrag, V. et al. Phase I/II study of MAK683 in patients with advanced malignancies, including diffuse large B-cell lymphoma. Blood 138, 1422 (2021).
Centore, R. C., Sandoval, G. J., Soares, L. M. M., Kadoch, C. & Chan, H. M. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950 (2020).
Ehrenhöfer-Wölfer, K. et al. SMARCA2-deficiency confers sensitivity to targeted inhibition of SMARCA4 in esophageal squamous cell carcinoma cell lines. Sci. Rep. 9, 11661 (2019).
Hoffman, G. R. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl Acad. Sci. USA 111, 3128–3133 (2014).
Schick, S. et al. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat. Genet. 51, 1399–1410 (2019).
Kofink, C. et al. A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo. Nat. Commun. 13, 5969 (2022).
Cantley, J. et al. Selective PROTAC-mediated degradation of SMARCA2 is efficacious in SMARCA4 mutant cancers. Nat. Commun. 13, 6814 (2022).
Hodges, C., Kirkland, J. G. & Crabtree, G. R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. https://doi.org/10.1101/cshperspect.a026930 (2016).
Michel, B. C. et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 20, 1410–1420 (2018).
Wang, X. et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017).
Li, J. et al. A role for SMARCB1 in synovial sarcomagenesis reveals that SS18–SSX induces canonical BAF destruction. Cancer Discov. 11, 2620–2637 (2021).
Brien, G. L. et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. eLife https://doi.org/10.7554/eLife.41305 (2018).
Jackson, K. L. et al. Abstract ND09: the discovery and characterization of CFT8634: a potent and selective degrader of BRD9 for the treatment of SMARCB1-perturbed cancers. Cancer Res. 82, ND09 (2022).
Milne, T. A. et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 (2002).
Mohan, M., Lin, C., Guest, E. & Shilatifard, A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer 10, 721–728 (2010).
Grembecka, J. et al. Menin–MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8, 277–284 (2012).
Daigle, S. R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Heikamp, E. B. et al. The menin–MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood 139, 894–906 (2022).
Uckelmann, H. J. et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science 367, 586–590 (2020).
Schübeler, D. et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 (2004).
Mohan, M. et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24, 574–589 (2010).
Min, J., Feng, Q., Li, Z., Zhang, Y. & Xu, R.-M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112, 711–723 (2003).
Sun, Y. et al. HOXA9 reprograms the enhancer landscape to promote leukemogenesis. Cancer Cell 34, 643–658 (2018).
Daigle, S. R. et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025 (2013).
Stein, E. M. et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131, 2661–2669 (2018).
Lin, J. et al. Menin ‘reads’ H3K79me2 mark in a nucleosomal context. Science 379, 717–723 (2023).
Yokoyama, A. et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123, 207–218 (2005).
Krivtsov, A. V. et al. A Menin–MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell 36, 660–673 (2019).
Shi, A. et al. Structural insights into inhibition of the bivalent menin–MLL interaction by small molecules in leukemia. Blood 120, 4461–4469 (2012).
Issa, G. C. et al. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature 615, 920–924 (2023).
Erba, H. P. et al. Update on a phase 1/2 first-in-human study of the menin–KMT2A (MLL) inhibitor ziftomenib (KO-539) in patients with relapsed or refractory acute myeloid leukemia. Blood 140, 153–156 (2022).
Malik, R. et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 21, 344–352 (2015).
Svoboda, L. K. et al. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget 8, 458–471 (2017).
Hemming, M. L. et al. MOZ and Menin–MLL complexes are complementary regulators of chromatin association and transcriptional output in gastrointestinal stromal tumor. Cancer Discov. 12, 1804–1823 (2022).
Perner, F. et al. MEN1 mutations mediate clinical resistance to menin inhibition. Nature 615, 913–919 (2023).
Lee, K. K. & Workman, J. L. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 (2007).
Yang, X.-J. & Seto, E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 (2008).
Welti, J. et al. Targeting the p300/CBP axis in lethal prostate cancer. Cancer Discov. 11, 1118–1137 (2021).
Nie, M. et al. Genome-wide CRISPR screens reveal synthetic lethal interaction between CREBBP and EP300 in diffuse large B-cell lymphoma. Cell Death Dis. 12, 419 (2021).
Durbin, A. D. et al. EP300 selectively controls the enhancer landscape of MYCN-amplified neuroblastoma. Cancer Discov. 12, 730–751 (2022).
Hay, D. A. et al. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains. J. Am. Chem. Soc. 136, 9308–9319 (2014).
Lasko, L. M. et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017).
Morrison-Smith, C. D. et al. Combined targeting of the BRD4–NUT–p300 axis in NUT midline carcinoma by dual selective bromodomain inhibitor, NEO2734. Mol. Cancer Ther. 19, 1406–1414 (2020).
Thomas, J. E. 2nd et al. Discovery of exceptionally potent, selective, and efficacious PROTAC degraders of CBP and p300 proteins. J. Med. Chem. 66, 8178–8199 (2023).
Vannam, R. et al. Targeted degradation of the enhancer lysine acetyltransferases CBP and p300. Cell Chem. Biol. 28, 503–514 (2021).
Borrow, J. et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 14, 33–41 (1996).
Yan, F. et al. KAT6A and ENL form an epigenetic transcriptional control module to drive critical leukemogenic gene-expression programs. Cancer Discov. 12, 792–811 (2022).
Northcott, P. A. et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 41, 465–472 (2009).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Turner-Ivey, B. et al. KAT6A, a chromatin modifier from the 8p11-p12 amplicon is a candidate oncogene in luminal breast cancer. Neoplasia 16, 644–655 (2014).
Saglam, O., Tang, Z., Tang, G., Medeiros, L. J. & Toruner, G. A. KAT6A amplifications are associated with shorter progression-free survival and overall survival in patients with endometrial serous carcinoma. PLoS ONE 15, e0238477 (2020).
Baell, J. B. et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257 (2018).
Lv, D. et al. Histone acetyltransferase KAT6A upregulates PI3K/AKT signaling through TRIM24 binding. Cancer Res. 77, 6190–6201 (2017).
Sharma, S. et al. Abstract 1130: first-in-class KAT6A/KAT6B inhibitor CTx-648 (PF-9363) demonstrates potent anti-tumor activity in ER+ breast cancer with KAT6A dysregulation. Cancer Res. 81, 1130 (2021).
Grant, P. A. et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900 (1999).
Jin, Q. et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011).
Farria, A. T. et al. Transcriptional activation of MYC-induced genes by GCN5 promotes B-cell lymphomagenesis. Cancer Res. 80, 5543–5553 (2020).
Mustachio, L. M., Roszik, J., Farria, A. & Dent, S. Y. R. Targeting the SAGA and ATAC transcriptional coactivator complexes in MYC-driven cancers. Cancer Res. 80, 1905–1911 (2020).
Bassi, Z. I. et al. Modulating PCAF/GCN5 immune cell function through a PROTAC approach. ACS Chem. Biol. 13, 2862–2867 (2018).
Fang, Y., Liao, G. & Yu, B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J. Hematol. Oncol. 12, 129 (2019).
Wang, Y. et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 (2009).
Kim, S.-A., Zhu, J., Yennawar, N., Eek, P. & Tan, S. Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate. Mol. Cell 78, 903–914 (2020).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Maes, T. et al. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell 33, 495–511 (2018).
Augert, A. et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci. Signal. https://doi.org/10.1126/scisignal.aau2922 (2019).
Mohammad, H. P. et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28, 57–69 (2015).
Sehrawat, A. et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc. Natl Acad. Sci. USA 115, E4179–E4188 (2018).
Salamero, O. et al. First-in-human phase I study of iadademstat (ORY-1001): a first-in-class lysine-specific histone demethylase 1A inhibitor, in relapsed or refractory acute myeloid leukemia. J. Clin. Oncol. 38, 4260–4273 (2020).
Bauer, T. M. et al. Phase I, open-label, dose-escalation study of the safety, pharmacokinetics, pharmacodynamics, and efficacy of GSK2879552 in relapsed/refractory SCLC. J. Thorac. Oncol. 14, 1828–1838 (2019).
McGinty, R. K., Henrici, R. C. & Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014).
Fursova, N. A. et al. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol. Cell 74, 1020–1036 (2019).
Parreno, V., Martinez, A.-M. & Cavalli, G. Mechanisms of Polycomb group protein function in cancer. Cell Res. 32, 231–253 (2022).
Scelfo, A. et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent- and RING1A/B-independent-specific activities. Mol. Cell 74, 1037–1052 (2019).
Schaefer, E. J. et al. BCOR and BCORL1 mutations drive epigenetic reprogramming and oncogenic signaling by unlinking PRC1.1 from target genes. Blood Cancer Discov. 3, 116–135 (2022).
Kang, J. H. et al. The mutation of BCOR is highly recurrent and oncogenic in mature T-cell lymphoma. BMC Cancer 21, 82 (2021).
Astolfi, A. et al. BCOR involvement in cancer. Epigenomics 11, 835–855 (2019).
Pisapia, D. J. et al. Fusions involving BCOR and CREBBP are rare events in infiltrating glioma. Acta Neuropathol. Commun. 8, 80 (2020).
Yamamoto, Y. et al. BCOR as a novel fusion partner of retinoic acid receptor α in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood 116, 4274–4283 (2010).
Tauziède-Espariat, A. et al. The EP300:BCOR fusion extends the genetic alteration spectrum defining the new tumoral entity of ‘CNS tumors with BCOR internal tandem duplication’. Acta Neuropathol. Commun. 8, 178 (2020).
Shukla, S. et al. Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain. Nat. Chem. Biol. 17, 784–793 (2021).
Rizo, A. et al. Repression of BMI1 in normal and leukemic human CD34+ cells impairs self-renewal and induces apoptosis. Blood 114, 1498–1505 (2009).
Maat, H. et al. The USP7–TRIM27 axis mediates non-canonical PRC1.1 function and is a druggable target in leukemia. iScience 24, 102435 (2021).
Wang, S. et al. A potent, selective CBX2 chromodomain ligand and its cellular activity during prostate cancer neuroendocrine differentiation. Chembiochem 22, 2335–2344 (2021).
Stuckey, J. I. et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat. Chem. Biol. 12, 180–187 (2016).
Milosevich, N. et al. Selective inhibition of CBX6: a methyllysine reader protein in the polycomb family. ACS Med. Chem. Lett. 7, 139–144 (2016).
Milosevich, N. et al. Polycomb paralog chromodomain inhibitors active against both CBX6 and CBX8*. ChemMedChem 16, 3027–3034 (2021).
Wang, S. et al. Optimization of ligands using focused DNA-encoded libraries to develop a selective, cell-permeable CBX8 chromodomain inhibitor. ACS Chem. Biol. 15, 112–131 (2020).
Erb, M. A. et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017).
Wan, L. et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269 (2017).
Hu, H. et al. The ENL YEATS epigenetic reader domain critically links MLL–ENL to leukemic stem cell frequency in t(11;19) leukemia. Leukemia 37, 190–201 (2023).
Perlman, E. J. et al. MLLT1 YEATS domain mutations in clinically distinctive favourable histology Wilms tumours. Nat. Commun. 6, 10013 (2015).
Wan, L. et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126 (2020).
Asiaban, J. N. et al. Cell-based ligand discovery for the ENL YEATS domain. ACS Chem. Biol. 15, 895–903 (2020).
Christott, T. et al. Discovery of a selective inhibitor for the YEATS domains of ENL/AF9. SLAS Discov. 24, 133–141 (2019).
Garnar-Wortzel, L. et al. Chemical inhibition of ENL/AF9 YEATS domains in acute leukemia. ACS Cent. Sci. 7, 815–830 (2021).
Jiang, Y. et al. Selective targeting of AF9 YEATS domain by cyclopeptide inhibitors with preorganized conformation. J. Am. Chem. Soc. 142, 21450–21459 (2020).
Li, X. et al. Structure-guided development of YEATS domain inhibitors by targeting π–π–π stacking. Nat. Chem. Biol. 14, 1140–1149 (2018).
Liu, Y. et al. Small-molecule inhibition of the acyl-lysine reader ENL as a strategy against acute myeloid leukemia. Cancer Discov. 12, 2684–2709 (2022).
Ma, X. R. et al. Discovery of selective small-molecule inhibitors for the ENL YEATS domain. J. Med. Chem. 64, 10997–11013 (2021).
Moustakim, M. et al. Discovery of an MLLT1/3 YEATS domain chemical probe. Angew. Chem. Int. Ed. Engl. 57, 16302–16307 (2018).
Li, X., Yao, Y., Wu, F. & Song, Y. A proteolysis-targeting chimera molecule selectively degrades ENL and inhibits malignant gene expression and tumor growth. J. Hematol. Oncol. 15, 41 (2022).
Pina, C., May, G., Soneji, S., Hong, D. & Enver, T. MLLT3 regulates early human erythroid and megakaryocytic cell fate. Cell Stem Cell 2, 264–273 (2008).
Gilan, O. et al. CRISPR–ChIP reveals selective regulation of H3K79me2 by Menin in MLL leukemia. Nat. Struct. Mol. Biol. 30, 1592–1606 (2023).
Zhu, L. et al. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 6, 770–783 (2016).
Aljazi, M. B., Gao, Y., Wu, Y., Mias, G. I. & He, J. Histone H3K36me2-specific methyltransferase ASH1L promotes MLL-AF9-induced leukemogenesis. Front. Oncol. 11, 754093 (2021).
Fujimoto, A. et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 48, 500–509 (2016).
Liu, L., Kimball, S., Liu, H., Holowatyj, A. & Yang, Z.-Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget 6, 2466–2482 (2015).
Xu, B. et al. Novel role of ASH1L histone methyltransferase in anaplastic thyroid carcinoma. J. Biol. Chem. 295, 8834–8845 (2020).
Rogawski, D. S. et al. Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity. Nat. Commun. 12, 2792 (2021).
Zhang, C., Xu, L., Zheng, X., Liu, S. & Che, F. Role of Ash1l in Tourette syndrome and other neurodevelopmental disorders. Dev. Neurobiol. 81, 79–91 (2021).
Walens, A. et al. Adaptation and selection shape clonal evolution of tumors during residual disease and recurrence. Nat. Commun. 11, 5017 (2020).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Gupta, P. B. et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644 (2011).
Shaffer, S. M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017).
Yang, D. et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell 185, 1905–1923 (2022).
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849 (2019).
Goyal, Y. et al. Diverse clonal fates emerge upon drug treatment of homogeneous cancer cells. Nature 620, 651–659 (2023).
Oren, Y. et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 596, 576–582 (2021).
Álvarez-Varela, A. et al. Mex3a marks drug-tolerant persister colorectal cancer cells that mediate relapse after chemotherapy. Nat. Cancer 3, 1052–1070 (2022).
Voigt, P., Tee, W.-W. & Reinberg, D. A double take on bivalent promoters. Genes Dev. 27, 1318–1338 (2013).
Marsolier, J. et al. H3K27me3 conditions chemotolerance in triple-negative breast cancer. Nat. Genet. 54, 459–468 (2022).
Mabe, N. W. et al. Epigenetic silencing of tumor suppressor Par-4 promotes chemoresistance in recurrent breast cancer. J. Clin. Invest. 128, 4413–4428 (2018).
Chaffer, C. L. et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 (2013).
Iniguez, A. B. et al. Resistance to epigenetic-targeted therapy engenders tumor cell vulnerabilities associated with enhancer remodeling. Cancer Cell 34, 922–938 (2018).
Ohta, Y. et al. Cell–matrix interface regulates dormancy in human colon cancer stem cells. Nature 608, 784–794 (2022).
Fane, M. E. et al. Stromal changes in the aged lung induce an emergence from melanoma dormancy. Nature 606, 396–405 (2022).
Warren, A. et al. Global computational alignment of tumor and cell line transcriptional profiles. Nat. Commun. 12, 22 (2021).
Murthy, P. K. L. et al. Epigenetic basis of oncogenic-Kras-mediated epithelial–cellular proliferation and plasticity. Dev. Cell 57, 310–328 (2022).
Filbin, M. G. et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335 (2018).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. Emt: 2016. Cell 166, 21–45 (2016).
Yang, J. et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
van Groningen, T. et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017).
Boeva, V. et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413 (2017).
Mabe, N. W. et al. Transition to a mesenchymal state in neuroblastoma confers resistance to anti-GD2 antibody via reduced expression of ST8SIA1. Nat. Cancer 3, 976–993 (2022).
Gartlgruber, M. et al. Super enhancers define regulatory subtypes and cell identity in neuroblastoma. Nat. Cancer 2, 114–128 (2021).
Linder, S. et al. Drug-induced epigenomic plasticity reprograms circadian rhythm regulation to drive prostate cancer toward androgen independence. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-21-0576 (2022).
Apfelbaum, A. A. et al. EWS::FLI1 and HOXD13 control tumor cell plasticity in Ewing sarcoma. Clin. Cancer Res. 28, 4466–4478 (2022).
Rubin, M. A., Bristow, R. G., Thienger, P. D., Dive, C. & Imielinski, M. Impact of lineage plasticity to and from a neuroendocrine phenotype on progression and response in prostate and lung cancers. Mol. Cell 80, 562–577 (2020).
Oser, M. G., Niederst, M. J., Sequist, L. V. & Engelman, J. A. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 16, e165–e172 (2015).
Hoek, K. S. et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 (2008).
Gardner, R. et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410 (2016).
Sáez-Ayala, M. et al. Directed phenotype switching as an effective antimelanoma strategy. Cancer Cell 24, 105–119 (2013).
Davies, A. et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 23, 1023–1034 (2021).
Yamamoto, S. et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell 25, 762–777 (2014).
Hinohara, K. et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 34, 939–953 (2018).
Drosos, Y. et al. NSD1 mediates antagonism between SWI/SNF and polycomb complexes and is required for transcriptional activation upon EZH2 inhibition. Mol. Cell 82, 2472–2489 (2022).
Sparbier, C. E. et al. Targeting Menin disrupts the KMT2A/B and polycomb balance to paradoxically activate bivalent genes. Nat. Cell Biol. 25, 258–272 (2023).
Zhang, Y. et al. Genome-wide CRISPR screen identifies PRC2 and KMT2D–COMPASS as regulators of distinct EMT trajectories that contribute differentially to metastasis. Nat. Cell Biol. 24, 554–564 (2022).
Avgustinova, A. et al. Loss of G9a preserves mutation patterns but increases chromatin accessibility, genomic instability and aggressiveness in skin tumours. Nat. Cell Biol. 20, 1400–1409 (2018).
Hogg, S. J. et al. Targeting histone acetylation dynamics and oncogenic transcription by catalytic P300/CBP inhibition. Mol. Cell 81, 2183–2200 (2021).
DuBois, S. G. et al. Randomized phase II trial of MIBG versus MIBG, vincristine, and irinotecan versus MIBG and vorinostat for patients with relapsed or refractory neuroblastoma: a report from NANT consortium. J. Clin. Oncol. 39, 3506–3514 (2021).
Gardner, E. E. et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2–SLFN11 axis. Cancer Cell 31, 286–299 (2017).
Kailayangiri, S. et al. EZH2 inhibition in Ewing sarcoma upregulates GD2 expression for targeting with gene-modified T cells. Mol. Ther. 27, 933–946 (2019).
Reppel, L. et al. Targeting disialoganglioside GD2 with chimeric antigen receptor-redirected T cells in lung cancer. J. Immunother. Cancer https://doi.org/10.1136/jitc-2021-003897 (2022).
Ennishi, D. et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 9, 546–563 (2019).
Palikyras, S. & Papantonis, A. Modes of phase separation affecting chromatin regulation. Open Biol. 9, 190167 (2019).
Lyon, A. S., Peeples, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
van der Lee, R. et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014).
Hansen, J. C., Lu, X., Ross, E. D. & Woody, R. W. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281, 1853–1856 (2006).
Kilgore, H. R. & Young, R. A. Learning the chemical grammar of biomolecular condensates. Nat. Chem. Biol. 18, 1298–1306 (2022).
Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020).
Chandra, B. et al. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov. 12, 1152–1169 (2022).
Tripathi, S. et al. Defining the condensate landscape of fusion oncoproteins. Nat. Commun. 14, 6008 (2023).
Chong, S. et al. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell 82, 2084–2097 (2022).
Seong, B. K. A. et al. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma. Cancer Cell 39, 1262–1278 (2021).
Chen, S. et al. Constitutive protein degradation induces acute cell death via proteolysis products. Preprint at bioRxiv https://doi.org/10.1101/2023.02.06.527237 (2023).
Belk, J. A., Daniel, B. & Satpathy, A. T. Epigenetic regulation of T cell exhaustion. Nat. Immunol. 23, 848–860 (2022).
Villanueva, L., Álvarez-Errico, D. & Esteller, M. The contribution of epigenetics to cancer immunotherapy. Trends Immunol. 41, 676–691 (2020).
Hiatt, J. B. et al. Inhibition of LSD1 with bomedemstat sensitizes small cell lung cancer to immune checkpoint blockade and T-cell killing. Clin. Cancer Res. 28, 4551–4564 (2022).
Acknowledgements
This work was funded by the National Institutes of Health (P01 CA217959 (K.S.), R35 CA283977 (K.S.), CA261035 (N.W.M.), CA279915 (N.W.M.), CA243266 (C.F.M.)), the Leukemia and Lymphoma Society, the Rally Foundation, and a Pediatric Stand Up to Cancer Catalyst Grant supported by Bristol-Myers Squibb (SU2C 6143). K.S. is also funded by the St. Jude Children’s Research Hospital Collaborative Research Program. We recognize that no review can be perfectly comprehensive. We apologize for any omissions of targets, drugs, or references due to the constraints of a Review article.
Author information
Authors and Affiliations
Contributions
N.W.M. was responsible for the writing of the manuscript, assembling of the figures and cataloging of clinical trial information tables. J.A.P. edited the manuscript and assisted in assembling clinical trial tables. C.F.M. wrote the section on histone acetyltransferases. K.S. supervised, edited and provided funding.
Corresponding author
Ethics declarations
Competing interests
K.S. receives grant funding from the DFCI/Novartis Drug Discovery Program and KronosBio, is a member of the scientific advisory board of and has stock options with Auron Therapeutics, and has consulted for AstraZeneca. C.F.M. is a current employee and has stock options at InduPro. All other authors have no disclosures.
Peer review
Peer review information
Nature Cancer thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mabe, N.W., Perry, J.A., Malone, C.F. et al. Pharmacological targeting of the cancer epigenome. Nat Cancer 5, 844–865 (2024). https://doi.org/10.1038/s43018-024-00777-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-024-00777-2
This article is cited by
-
ER-phagy mediates the anti-tumoral synergism between HDAC inhibition and chemotherapy
Cell Communication and Signaling (2025)
-
The ESRP1 promoter reporter can function as an in vivo sensor of DNA methyltransferase inhibition
BMC Biotechnology (2025)
-
EZH2 inhibitor SHR2554 enhances the anti-tumor efficacy of HDAC inhibitor Chidamide through STAT1 in T-cell lymphoma
Cell Death & Disease (2025)
-
Getting the right combination to break the epigenetic code
Nature Reviews Clinical Oncology (2025)
-
Valproic acid improves the efficacy of oxaliplatin/fluoropyrimidine-based chemotherapy by targeting cancer stem cell via β-Catenin modulation in colorectal cancer
Cell Death & Disease (2025)