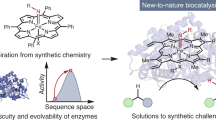
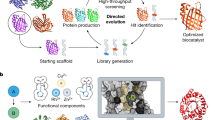
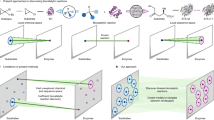
Abstract
Biocatalysis has become an important aspect of modern organic synthesis, both in academia and across the chemical and pharmaceutical industries. Its success has been largely due to a rapid expansion of the range of chemical reactions accessible, made possible by advanced tools for enzyme discovery coupled with high-throughput laboratory evolution techniques for biocatalyst optimization. A wide range of tailor-made enzymes with high efficiencies and selectivities can now be produced quickly and on a gram to kilogram scale, with dedicated databases and search tools aimed at making these biocatalysts accessible to a broader scientific community. This Primer discusses the current state-of-the-art methodology in the field, including route design, enzyme discovery, protein engineering and the implementation of biocatalysis in industry. We highlight recent advances, such as de novo design and directed evolution, and discuss parameters that make a good reproducible biocatalytic process for industry. The general concepts will be illustrated by recent examples of applications in academia and industry, including the development of multistep enzyme cascades.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 1 digital issues and online access to articles
$119.00 per year
only $119.00 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Yamada, H. & Kobayashi, M. Nitrile hydratase and its application to industrial production of acrylamide. Biosci. Biotechnol. Biochem. 60, 1391–1400 (1996).
Kirk, O., Borchert, T. V. & Fuglsang, C. C. Industrial enzyme applications. Curr. Opin. Biotechnol. 13, 345–351 (2002).
Devine, P. N. et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2, 409–421 (2018).
Wu, S., Snajdrova, R., Moore, J. C., Baldenius, K. & Bornscheuer, U. T. Biocatalysis: enzymatic synthesis for industrial applications. Angew. Chem. Int. Ed. 60, 88–119 (2021).
Sheldon, R. A., Brady, D. & Bode, M. L. The hitchhiker’s guide to biocatalysis: recent advances in the use of enzymes in organic synthesis. Chem. Sci. 11, 2587–2605 (2020). This article presents an excellent recent overall review of biocatalysis.
Birmingham, W. R. et al. Bioretrosynthetic construction of a didanosine biosynthetic pathway. Nat. Chem. Biol. 10, 392–399 (2014). This article develops the concept of ‘bio-retrosynthesis’ and its application to afford an important biomolecule.
Huffman, M. A. et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 366, 1255–1259 (2019). This article presents the very impressive development of a multistep enzyme cascade with multiple enzyme engineering challenges by an industrial team.
Finnigan, W. et al. RetroBioCat as a computer-aided synthesis planning tool for biocatalytic reactions and cascades. Nat. Catal. 4, 98–104 (2021). This article establishes a database and computer-aided synthesis planning tool that allows scientists to design enzyme cascades.
Charnock, S., Bernardini, A. M., Monza, E., Lucas, M. F. & Sutton, P. W. in Applied Biocatalysis (eds Whittall, J. & Sutton, P. W.) 27–133 (Wiley, 2020).
Bateman, A. et al. UniProt: the Universal Protein Knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017).
Buchholz, P. C. F. et al. BioCatNet: a database system for the integration of enzyme sequences and biocatalytic experiments. ChemBioChem 17, 2093–2098 (2016).
Scott, T. A. & Piel, J. The hidden enzymology of bacterial natural product biosynthesis. Nat. Rev. Chem. 3, 404–425 (2019).
Martinez, S. & Hausinger, R. P. Catalytic mechanisms of Fe(II)- and 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 290, 20702–20711 (2015).
Zwick, C. R. & Renata, H. Harnessing the biocatalytic potential of iron- and α-ketoglutarate-dependent dioxygenases in natural product total synthesis. Nat. Prod. Rep. 37, 1065–1079 (2020).
Lukat, P. et al. Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity. Chem. Sci. 8, 7521–7527 (2017).
Zwick, C. R. & Renata, H. Remote C–H hydroxylation by an α-ketoglutarate-dependent dioxygenase enables efficient chemoenzymatic synthesis of manzacidin C and proline analogs. J. Am. Chem. Soc. 140, 1165–1169 (2018).
Neugebauer, M. E. et al. A family of radical halogenases for the engineering of amino-acid-based products. Nat. Chem. Biol. 15, 1009–1016 (2019).
Chekan, J. R. et al. Scalable biosynthesis of the seaweed neurochemical, kainic acid. Angew. Chem. Int. Ed. 58, 8454–8457 (2019).
Chakrabarty, S., Wang, Y., Perkins, J. C. & Narayan, A. R. H. Scalable biocatalytic C–H oxyfunctionalization reactions. Chem. Soc. Rev. 49, 8137–8155 (2020). This article presents an excellent review on the current state of the art of C–H oxyfunctionalizations of organic molecules using biocatalysis.
Liao, C. & Seebeck, F. P. S-Adenosylhomocysteine as a methyl transfer catalyst in biocatalytic methylation reactions. Nat. Catal. 2, 696–701 (2019).
Marsh, C. O. et al. A natural Diels–Alder biocatalyst enables efficient [4 + 2] cycloaddition under harsh reaction conditions. ChemCatChem 11, 5027–5031 (2019).
Fisher, B. F., Snodgrass, H. M., Jones, K. A., Andorfer, M. C. & Lewis, J. C. Site-selective C–H halogenation using flavin-dependent halogenases identified via family-wide activity profiling. ACS Cent. Sci. 5, 1844–1856 (2019).
Galanie, S., Entwistle, D. & Lalonde, J. Engineering biosynthetic enzymes for industrial natural product synthesis. Nat. Prod. Rep. 37, 1122–1143 (2020).
Schultz, B. J., Kim, S. Y., Lau, W. & Sattely, E. S. Total biosynthesis for milligram-scale production of etoposide intermediates in a plant chassis. J. Am. Chem. Soc. 141, 19231–19235 (2019).
Chen, M., Liu, C. T. & Tang, Y. Discovery and biocatalytic application of a PLP-dependent amino acid γ-substitution enzyme that catalyzes C–C bond formation. J. Am. Chem. Soc. 142, 10506–10515 (2020).
Bar-Even, A. et al. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 (2011).
Goldsmith, M. & Tawfik, D. S. Enzyme engineering: reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 47, 140–150 (2017).
Handelsman, J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685 (2004).
Iqbal, H. A., Feng, Z. & Brady, S. F. Biocatalysts and small molecule products from metagenomic studies. Curr. Opin. Chem. Biol. 16, 109–116 (2012).
Coscolín, C. et al. Bioprospecting reveals class ω-transaminases converting bulky ketones and environmentally relevant polyamines. Appl. Environ. Microbiol. 85, e02404-18 (2019).
Green, A. P., Turner, N. J. & O’Reilly, E. Chiral amine synthesis using ω-transaminases: an amine donor that displaces equilibria and enables high-throughput screening. Angew. Chem. Int. Ed. 53, 10714–10717 (2014).
Baud, D., Ladkau, N., Moody, T. S., Ward, J. M. & Hailes, H. C. A rapid, sensitive colorimetric assay for the high-throughput screening of transaminases in liquid or solid-phase. Chem. Commun. 51, 17225–17228 (2015).
Nasseri, S. A., Betschart, L., Opaleva, D., Rahfeld, P. & Withers, S. G. A mechanism-based approach to screening metagenomic libraries for discovery of unconventional glycosidases. Angew. Chem. Int. Ed. 57, 11359–11364 (2018).
Colin, P. Y. et al. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 6, 1–12 (2015).
Rahfeld, P. et al. An enzymatic pathway in the human gut microbiome that converts A to universal O type blood. Nat. Microbiol. 4, 1475–1485 (2019). This article identifies a two-enzyme system from a large gut microbiome library that enables generation of universal O-type blood from A-type donors.
Smith, D. R. M. et al. An unusual flavin-dependent halogenase from the metagenome of the marine sponge Theonella swinhoei WA. ACS Chem. Biol. 12, 1281–1287 (2017).
Baud, D., Jeffries, J. W. E., Moody, T. S., Ward, J. M. & Hailes, H. C. A metagenomics approach for new biocatalyst discovery: application to transaminases and the synthesis of allylic amines. Green. Chem. 19, 1134–1143 (2017).
Armstrong, Z. et al. Metagenomics reveals functional synergy and novel polysaccharide utilization loci in the Castor canadensis fecal microbiome. ISME J. 12, 2757–2769 (2018).
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. 57, 4143–4148 (2018). This article presents a review of directed evolution by the 2018 Nobel Prize Laureate for Chemistry.
Qu, G., Li, A., Acevedo-Rocha, C. G., Sun, Z. & Reetz, M. T. The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chem. Int. Ed. 59, 13204–13231 (2020).
Reetz, M. T., Bocola, M., Carballeira, J. D., Zha, D. & Vogel, A. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem. Int. Ed. 44, 4192–4196 (2005).
Reetz, M. T., Kahakeaw, D. & Lohmer, R. Addressing the numbers problem in directed evolution. ChemBioChem 9, 1797–1804 (2008).
Kille, S. et al. Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth. Biol. 2, 83–92 (2013).
Cadwell, R. C. & Joyce, G. F. Mutagenic PCR. CSH Protoc. https://doi.org/10.1101/pdb.prot4143 (2006).
Stemmer, W. P. C. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994). This article establishes DNA shuffling as a method for generation of gene libraries for directed evolution.
Schober, M. et al. Chiral synthesis of LSD1 inhibitor GSK2879552 enabled by directed evolution of an imine reductase. Nat. Catal. 2, 909–915 (2019).
Heath, R. S., Pontini, M., Bechi, B. & Turner, N. J. Development of an R-selective amine oxidase with broad substrate specificity and high enantioselectivity. ChemCatChem 6, 996–1002 (2014).
Weiß, M. S., Pavlidis, I. V., Vickers, C., Hohne, M. & Bornscheuer, U. T. Glycine oxidase based high-throughput solid-phase assay for substrate profiling and directed evolution of (R)- and (S)-selective amine transaminases. Anal. Chem. 86, 11847–11853 (2014).
Yan, C. et al. Real-time screening of biocatalysts in live bacterial colonies. J. Am. Chem. Soc. 139, 1408–1411 (2017). This article presents recent approaches to using label-free screening methods based on mass spectrometry.
Becker, S., Schmoldt, H. U., Adams, T. M., Wilhelm, S. & Kolmar, H. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts. Curr. Opin. Biotechnol. 15, 323–329 (2004).
Chen, T. et al. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2′-modified DNA. Nat. Chem. 8, 556–562 (2016).
Agresti, J. J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl Acad. Sci. USA 107, 4004–4009 (2010).
Obexer, R. et al. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat. Chem. 9, 50–56 (2017).
Wang, L. & Schultz, P. G. A general approach for the generation of orthogonal tRNAs. Chem. Biol. 8, 883–890 (2001).
Bryson, D. I. et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 13, 1253–1260 (2017).
Ravikumar, A., Arrieta, A. & Liu, C. C. An orthogonal DNA replication system in yeast. Nat. Chem. Biol. 10, 175–177 (2014).
Ghislieri, D. et al. Engineering an enantioselective amine oxidase for the synthesis of pharmaceutical building blocks and alkaloid natural products. J. Am. Chem. Soc. 135, 10863–10869 (2013).
Debon, A. et al. Ultrahigh-throughput screening enables efficient single-round oxidase remodelling. Nat. Catal. 2, 740–747 (2019).
Holland-Moritz, D. A. et al. Mass activated droplet sorting (MADS) enables high-throughput screening of enzymatic reactions at nanoliter scale. Angew. Chem. Int. Ed. 59, 4470–4477 (2020). This article applies droplet sorting as one of the most successful methods for ultra-high-throughput screening of enzyme libraries.
Wan, W., Tharp, J. M. & Liu, W. R. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta 1844, 1059–1070 (2014).
Warshel, A. et al. Electrostatic basis for enzyme catalysis. Chem. Rev. 106, 3210–3235 (2006).
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Osuna, S. The challenge of predicting distal active site mutations in computational enzyme design. WIREs Comput. Mol. Sci. 11, e1502 (2021).
Kiss, G., Çelebi-Ölçüm, N., Moretti, R., Baker, D. & Houk, K. N. Computational enzyme design. Angew. Chem. Int. Ed. 52, 5700–5725 (2013).
Privett, H. K. et al. Iterative approach to computational enzyme design. Proc. Natl Acad. Sci. USA 109, 3790–3795 (2012).
Wijma, H. J. et al. Enantioselective enzymes by computational design and in silico screening. Angew. Chem. 127, 3797–3801 (2015).
Davey, J. A. & Chica, R. A. Multistate approaches in computational protein design. Protein Sci. 21, 1241–1252 (2012).
Mondal, D., Kolev, V. & Warshel, A. Combinatorial approach for exploring conformational space and activation barriers in computer-aided enzyme design. ACS Catal. 10, 6002–6012 (2020).
Maria-Solano, M. A., Serrano-Hervás, E., Romero-Rivera, A., Iglesias-Fernández, J. & Osuna, S. Role of conformational dynamics in the evolution of novel enzyme function. Chem. Commun. 54, 6622–6634 (2018).
Campbell, E. C. et al. Laboratory evolution of protein conformational dynamics. Curr. Opin. Struct. Biol. 50, 49–57 (2018).
Crean, R. M., Gardner, J. M. & Kamerlin, S. C. L. Harnessing conformational plasticity to generate designer enzymes. J. Am. Chem. Soc. 142, 11324–11342 (2020).
Kreß, N., Halder, J. M., Rapp, L. R. & Hauer, B. Unlocked potential of dynamic elements in protein structures: channels and loops. Curr. Opin. Chem. Biol. 47, 109–116 (2018).
Vavra, O. et al. CaverDock: a molecular docking-based tool to analyse ligand transport through protein tunnels and channels. Bioinformatics 35, 4986–4993 (2019).
Otten, R. et al. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 370, 1442–1446 (2020).
Romero-Rivera, A., Garcia-Borràs, M. & Osuna, S. Role of conformational dynamics in the evolution of retro-aldolase activity. ACS Catal. 7, 8524–8532 (2017).
Casini, A. et al. A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J. Am. Chem. Soc. 140, 4302–4316 (2018).
Gardner, J. M., Biler, M., Risso, V. A., Sanchez-Ruiz, J. M. & Kamerlin, S. C. L. Manipulating conformational dynamics to repurpose ancient proteins for modern catalytic functions. ACS Catal. 10, 4863–4870 (2020).
Pabis, A., Risso, V. A., Sanchez-Ruiz, J. M. & Kamerlin, S. C. Cooperativity and flexibility in enzyme evolution. Curr. Opin. Struct. Biol. 48, 83–92 (2018).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010). This article reviews the methods to expand the genetic code for the introduction of non-natural amino acids into proteins.
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Seyedsayamdost, M. R., Xie, J., Chan, C. T. Y., Schultz, P. G. & Stubbe, J. Site-specific insertion of 3-aminotyrosine into subunit α2 of E. coli ribonucleotide reductase: direct evidence for involvement of Y730 and Y731 in radical propagation. J. Am. Chem. Soc. 129, 15060–15071 (2007).
Faraldos, J. A. et al. Probing eudesmane cation–π interactions in catalysis by aristolochene synthase with non-canonical amino acids. J. Am. Chem. Soc. 133, 13906–13909 (2011).
Wu, Y. & Boxer, S. G. A critical test of the electrostatic contribution to catalysis with noncanonical amino acids in ketosteroid isomerase. J. Am. Chem. Soc. 138, 11890–11895 (2016).
Huguenin-Dezot, N. et al. Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid. Nature 565, 112–117 (2019).
Ortmayer, M. et al. Rewiring the ‘push–pull’ catalytic machinery of a heme enzyme using an expanded genetic code. ACS Catal. 10, 2735–2746 (2020).
Burke, A. J. et al. Design and evolution of an enzyme with a non-canonical organocatalytic mechanism. Nature 570, 219–223 (2019). This article is a recent example of using non-canonical amino acids in protein evolution to obtain new enzyme activities.
Drienovská, I., Mayer, C., Dulson, C. & Roelfes, G. A designer enzyme for hydrazone and oxime formation featuring an unnatural catalytic aniline residue. Nat. Chem. 10, 946–952 (2018).
Santoro, S. W., Wang, L., Herberich, B., King, D. S. & Schultz, P. G. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 20, 1044–1048 (2002).
Pott, M. et al. A noncanonical proximal heme ligand affords an efficient peroxidase in a globin fold. J. Am. Chem. Soc. 140, 1535–1543 (2018).
Li, J. C., Liu, T., Wang, Y., Mehta, A. P. & Schultz, P. G. Enhancing protein stability with genetically encoded noncanonical amino acids. J. Am. Chem. Soc. 140, 15997–16000 (2018).
Wurz, R. P. Chiral dialkylaminopyridine catalysts in asymmetric synthesis. Chem. Rev. 107, 5570–5595 (2007).
Bolon, D. N. & Mayo, S. L. Enzyme-like proteins by computational design. Proc. Natl Acad. Sci. USA 98, 14274–14279 (2001).
Richter, F. et al. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. J. Am. Chem. Soc. 134, 16197–16206 (2012).
Moroz, Y. S. et al. New tricks for old proteins: single mutations in a nonenzymatic protein give rise to various enzymatic activities. J. Am. Chem. Soc. 137, 14905–14911 (2015).
Burton, A. J., Thomson, A. R., Dawson, W. M., Brady, R. L. & Woolfson, D. N. Installing hydrolytic activity into a completely de novo protein framework. Nat. Chem. 8, 837–844 (2016).
Nazor, J., Liu, J. & Huisman, G. Enzyme evolution for industrial biocatalytic cascades. Curr. Opin. Biotechnol. 69, 182–190 (2021). This review focuses on recent industrial biocatalytic cascades.
McIntosh, J. A. & Owens, A. Enzyme engineering for biosynthetic cascades. Curr. Opin. Green Sustain. Chem. 29, 100448 (2021).
Schrittwieser, J. H., Velikogne, S., Hall, M. & Kroutil, W. Artificial biocatalytic linear cascades for preparation of organic molecules. Chem. Rev. 118, 270–348 (2018).
Mayer, S. F., Kroutil, W. & Faber, K. Enzyme-initiated domino (cascade) reactions. Chem. Soc. Rev. 30, 332–339 (2001).
García-Junceda, E., Lavandera, I., Rother, D. & Schrittwieser, J. H. (Chemo)enzymatic cascades — nature’s synthetic strategy transferred to the laboratory. J. Mol. Catal. B Enzym. 114, 1–6 (2015).
Rudroff, F. et al. Opportunities and challenges for combining chemo- and biocatalysis. Nat. Catal. 1, 12–22 (2018).
France, S. P., Hepworth, L. J., Turner, N. J. & Flitsch, S. L. Constructing biocatalytic cascades: in vitro and in vivo approaches to de novo multi-enzyme pathways. ACS Catal. 7, 710–724 (2017). This article reviews the literature on a wide range of multienzyme de novo cascades using isolated enzymes and whole-cell systems.
Turner, N. J. & O’Reilly, E. Biocatalytic retrosynthesis. Nat. Chem. Biol. 9, 285–288 (2013). This review develops the concept of biocatalytic retrosynthesis.
Hönig, M., Sondermann, P., Turner, N. J. & Carreira, E. M. Enantioselective chemo- and biocatalysis: partners in retrosynthesis. Angew. Chem. Int. Ed. 56, 8942–8973 (2017).
Green, A. P. & Turner, N. J. Biocatalytic retrosynthesis: redesigning synthetic routes to high-value chemicals. Perspect. Sci. 9, 42–48 (2016).
de Souza, R. O. M. A., Miranda, L. S. M. & Bornscheuer, U. T. A retrosynthesis approach for biocatalysis in organic synthesis. Chem. A Eur. J. 23, 12040–12063 (2017).
Bachmann, B. O. Biosynthesis: is it time to go retro? Nat. Chem. Biol. 6, 390–393 (2010).
Hadadi, N. & Hatzimanikatis, V. Design of computational retrobiosynthesis tools for the design of de novo synthetic pathways. Curr. Opin. Chem. Biol. 28, 99–104 (2015).
Coley, C. W., Green, W. H. & Jensen, K. F. Machine learning in computer-aided synthesis planning. Acc. Chem. Res. 51, 1281–1289 (2018).
Mohamad, N. R., Marzuki, N. H. C., Buang, N. A., Huyop, F. & Wahab, R. A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 29, 205–220 (2015).
Basso, A. & Serban, S. Industrial applications of immobilized enzymes — a review. Mol. Catal. 479, 110607 (2019).
Fryszkowska, A. & Devine, P. N. Biocatalysis in drug discovery and development. Curr. Opin. Chem. Biol. 55, 151–160 (2020).
Latham, J. et al. in Applied Biocatalysis (eds Whittall, J. & Sutton, P. W.) 1–25 (Wiley, 2020).
Prier, C. K. & Kosjek, B. Recent preparative applications of redox enzymes. Curr. Opin. Chem. Biol. 49, 105–112 (2019).
Woodley, J. M. New frontiers in biocatalysis for sustainable synthesis. Curr. Opin. Green. Sustain. Chem. 21, 22–26 (2020).
Sheldon, R. A. & Woodley, J. M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 118, 801–838 (2018).
Sheldon, R. A. Metrics of green chemistry and sustainability: past, present, and future. ACS Sustain. Chem. Eng. 6, 32–48 (2018).
Tieves, F. et al. Energising the E-factor: the E+-factor. Tetrahedron 75, 1311–1314 (2019).
Hinzmann, A., Glinski, S., Worm, M. & Gröger, H. Enzymatic synthesis of aliphatic nitriles at a substrate loading of up to 1.4 kg/l: a biocatalytic record achieved with a heme protein. J. Org. Chem. 84, 4867–4872 (2019).
Bornadel, A. et al. Technical considerations for scale-up of imine-reductase-catalyzed reductive amination: a case study. Org. Process. Res. Dev. 23, 1262–1268 (2019).
Hülsewede, D., Meyer, L. & von Langermann, J. Application of in situ product crystallization and related techniques in biocatalytic processes. Chem. A Eur. J. 25, 4871–4884 (2019).
Fellechner, O., Blatkiewicz, M. & Smirnova, I. Reactive separations for in situ product removal of enzymatic reactions: a review. Chem. Ing. Tech. 91, 1522–1543 (2019).
Aalbers, F. S. et al. Approaching boiling point stability of an alcohol dehydrogenase through computationally-guided enzyme engineering. eLife 9, e54639 (2020).
Gumulya, Y. et al. Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nat. Catal. 1, 878–888 (2018).
Thomas, A., Cutlan, R., Finnigan, W., van der Giezen, M. & Harmer, N. Highly thermostable carboxylic acid reductases generated by ancestral sequence reconstruction. Commun. Biol. 2, 1–12 (2019).
Nicoll, C. R. et al. Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs. Nat. Struct. Mol. Biol. 27, 14–24 (2020).
Gomez-Fernandez, B. J., Risso, V. A., Rueda, A., Sanchez-Ruiz, J. M. & Alcalde, M. Ancestral resurrection and directed evolution of fungal mesozoic laccases. Appl. Environ. Microbiol. 86, e00778-20 (2020).
Carletti, M. S. et al. Revenant: a database of resurrected proteins. Database. 2020, 31 (2020).
Truppo, M. D., Strotman, H. & Hughes, G. Development of an immobilized transaminase capable of operating in organic solvent. ChemCatChem 4, 1071–1074 (2012).
Mattey, A. P. et al. Natural heterogeneous catalysis with immobilised oxidase biocatalysts. RSC Adv. 10, 19501–19505 (2020).
Böhmer, W. et al. Highly efficient production of chiral amines in batch and continuous flow by immobilized ω-transaminases on controlled porosity glass metal-ion affinity carrier. J. Biotechnol. 291, 52–60 (2019).
Britton, J., Majumdar, S. & Weiss, G. A. Continuous flow biocatalysis. Chem. Soc. Rev. 47, 5891–5918 (2018).
Rodrigues, R. C., Ortiz, C., Berenguer-Murcia, Á., Torres, R. & Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 42, 6290–6307 (2013).
Sheldon, R. A. in Green Biocatalysis (ed. Patel, R. N.) 1–15 (Wiley, 2016).
Cespugli, M. et al. Rice husk as an inexpensive renewable immobilization carrier for biocatalysts employed in the food, cosmetic and polymer sectors. Catalysts 8, 471 (2018).
Woodley, J. M. Towards the sustainable production of bulk-chemicals using biotechnology. N. Biotechnol. 59, 59–64 (2020).
Hughes, D. L. Biocatalysis in drug development — highlights of the recent patent literature. Org. Process. Res. Dev. 22, 1063–1080 (2018).
de María, P., de Gonzalo, G. & Alcántara, A. Biocatalysis as useful tool in asymmetric synthesis: an assessment of recently granted patents (2014–2019). Catalysts 9, 802 (2019).
Hauer, B. Embracing nature’s catalysts: a viewpoint on the future of biocatalysis. ACS Catal. 10, 8418–8427 (2020). This article presents an insightful review of the future challenges of biocatalysis in academia and industry.
Jiao, S., Li, F., Yu, H. & Shen, Z. Advances in acrylamide bioproduction catalyzed with Rhodococcus cells harboring nitrile hydratase. Appl. Microbiol. Biotechnol. 104, 1001–1012 (2020).
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010).
Eichhorn, E. et al. Biocatalytic process for (−)-ambrox production using squalene hopene cyclase. Adv. Synth. Catal. 360, 2339–2351 (2018).
Baker Dockrey, S. A., Lukowski, A. L., Becker, M. R. & Narayan, A. R. H. Biocatalytic site- and enantioselective oxidative dearomatization of phenols. Nat. Chem. 10, 119–125 (2018).
Li, G., Wang, J.-B. & Reetz, M. T. Biocatalysts for the pharmaceutical industry created by structure-guided directed evolution of stereoselective enzymes. Bioorg. Med. Chem. 26, 1241–1251 (2018).
Arora, K. K. et al. Manufacturing Process and Intermediates for a Pyrrolo[2,3- D]Pyrimidine Compound and use Thereof. US Patent 10,815,240 (2020).
Jaeger, K. E., Eggert, T., Eipper, A. & Reetz, M. T. Directed evolution and the creation of enantioselective biocatalysts. Appl. Microbiol. Biotechnol. 55, 519–530 (2001).
Manning, J. et al. Regio- and enantio-selective chemo-enzymatic C–H-lactonization of decanoic acid to (S)-δ-decalactone. Angew. Chem. Int. Ed. 58, 5668–5671 (2019).
Zhang, J. et al. Engineered C–N lyase: enantioselective synthesis of chiral synthons for artificial dipeptide sweeteners. Angew. Chem. 132, 437–443 (2020).
Bruggink, A., Schoevaart, R. & Kieboom, T. Concepts of nature in organic synthesis: cascade catalysis and multistep conversions in concert. Org. Process. Res. Dev. 7, 622–640 (2003).
Sperl, J. M. & Sieber, V. Multienzyme cascade reactions — status and recent advances. ACS Catal. 8, 2385–2396 (2018).
Lenz, M., Borlinghaus, N., Weinmann, L. & Nestl, B. M. Recent advances in imine reductase-catalyzed reactions. World J. Microbiol. Biotechnol. 33, 199 (2017).
Schaffer, S. et al. Producing amines and diamines from a carboxylic acid or dicarboxylic acid or a monoester thereof. US Patent 9,725,746 (2017)
Benkovics, T. et al. Evolving to an ideal synthesis of molnupiravir, an investigational treatment for COVID-19. Preprint at https://doi.org/10.26434/chemrxiv.13472373.v1 (2020).
Yin, Z. et al. Computing platforms for big biological data analytics: perspectives and challenges. Comput. Struct. Biotechnol. J. 15, 403–411 (2017).
Agarwala, R. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44, D7–D19 (2016).
Goodman, J. Computer sSoftware review: Reaxys Reaxys. Elsevier Properties SA 360 Park Avenue South, New York, NY 10010-1710. www.info.reaxys.com. J. Chem. Inf. Model. 49, 2897–2898 (2009).
Garritano, J. R. Evolution of SciFinder, 2011–2013: new features, new content. Sci. Technol. Libr. 32, 346–371 (2013).
Jeske, L., Placzek, S., Schomburg, I., Chang, A. & Schomburg, D. BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res. 47, D542–D549 (2019).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Gunera, J., Kindinger, F., Li, S. M. & Kolb, P. PrenDB, a substrate prediction database to enable biocatalytic use of prenyltransferases. J. Biol. Chem. 292, 4003–4021 (2017).
Van Santen, J. A. et al. The natural products atlas: an open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 5, 1824–1833 (2019).
Tipton, K. F. et al. Standards for reporting enzyme data: the STRENDA consortium: what it aims to do and why it should be helpful. Perspect. Sci. 1, 131–137 (2014).
Swainston, N. et al. STRENDA DB: enabling the validation and sharing of enzyme kinetics data. FEBS J. 285, 2193–2204 (2018).
Perkel, J. M. The Internet of Things comes to the lab. Nature 542, 125–126 (2017).
Jennings-Antipov, L. D. & Gardner, T. S. Digital publishing isn’t enough: the case for ‘blueprints’ in scientific communication. Emerg. Top. Life Sci. 2, 755–758 (2018).
Goodwin, N. C., Morrison, J. P., Fuerst, D. E. & Hadi, T. Biocatalysis in medicinal chemistry: challenges to access and drivers for adoption. ACS Med. Chem. Lett. 10, 1363–1366 (2019).
Nielsen, J. & Keasling, J. D. Engineering cellular metabolism. Cell 164, 1185–1197 (2016).
Truppo, M. D. Biocatalysis in the pharmaceutical industry: the need for speed. ACS Med. Chem. Lett. 8, 476–480 (2017). This article presents an excellent review of the adaptation and challenges of biocatalysis in the pharmaceutical industry, with particular focus on timescales of process development.
Yang, K. K., Wu, Z. & Arnold, F. H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 16, 687–694 (2019).
Markel, U. et al. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 49, 233–262 (2020).
Bunzel, H. A., Garrabou, X., Pott, M. & Hilvert, D. Speeding up enzyme discovery and engineering with ultrahigh-throughput methods. Curr. Opin. Struct. Biol. 48, 149–156 (2018).
Hillson, N. et al. Building a global alliance of biofoundries. Nat. Commun. 10, 1–4 (2019).
Windle, C. L., Müller, M., Nelson, A. & Berry, A. Engineering aldolases as biocatalysts. Curr. Opin. Chem. Biol. 19, 25–33 (2014).
Brovetto, M., Gamenara, D., Saenz Méndez, P. & Seoane, G. A. C–C bond-forming lyases in organic synthesis. Chem. Rev. 111, 4346–4403 (2011).
Zetzsche, L. E. & Narayan, A. R. H. Broadening the scope of biocatalytic C–C bond formation. Nat. Rev. Chem. 4, 334–346 (2020).
Li, Z. et al. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 295, 833–849 (2020).
Fessner, N. D. P450 monooxygenases enable rapid late-stage diversification of natural products via C–H bond activation. ChemCatChem 11, 2226–2242 (2019).
Lall, M. S. et al. Late-stage lead diversification coupled with quantitative nuclear magnetic resonance spectroscopy to identify new structure–activity relationship vectors at nanomole-scale synthesis: application to loratadine, a human histamine H1 receptor inverse agonist. J. Med. Chem. 63, 7268–7292 (2020).
Dong, J. J. et al. Biocatalytic oxidation reactions: a chemist’s perspective. Angew. Chem. Int. Ed. 57, 9238–9261 (2018).
Silverman, A. D., Karim, A. S. & Jewett, M. C. Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21, 151–170 (2020).
Khambhati, K. et al. Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems. Front. Bioeng. Biotechnol. 7, 248 (2019).
Zimmerman, J. B., Anastas, P. T., Erythropel, H. C. & Leitner, W. Designing for a green chemistry future. Science 367, 397–400 (2020).
Hammer, S. C., Knight, A. M. & Arnold, F. H. Design and evolution of enzymes for non-natural chemistry. Curr. Opin. Green Sustain. Chem. 7, 23–30 (2017).
DeHovitz, J. S. et al. Static to inducibly dynamic stereocontrol: the convergent use of racemic β-substituted ketones. Science 369, 1113–1118 (2020).
O’Reilly, E., Köhler, V., Flitsch, S. L. & Turner, N. J. Cytochromes P450 as useful biocatalysts: addressing the limitations. Chem. Commun. 47, 2490–2501 (2011).
Basler, S. et al. Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold. Nat. Chem. 13, 231–235 (2021).
Liu, Z. & Arnold, F. H. New-to-nature chemistry from old protein machinery: carbene and nitrene transferases. Curr. Opin. Biotechnol. 69, 43–51 (2021). This review describes recent developments of new to nature reactions catalysed by engineered enzymes.
Callaway, E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 588, 203–204 (2020). This article presents a recent breakthrough in computational protein structure prediction from the primary sequence.
Dou, J. et al. De novo design of a fluorescence-activating β-barrel. Nature 561, 485–491 (2018).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
Huang, P. S., Boyken, S. E. & Baker, D. The coming of age of de novo protein design. Nature 537, 320–327 (2016).
Fischer, M. & Pleiss, J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 31, 319–321 (2003).
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). This article discusses the curated comprehensive database of carbohydrate-active enzymes (CAZymes) that has been a useful tool to the glycoscience community.
Savelli, B. et al. RedoxiBase: a database for ROS homeostasis regulated proteins. Redox Biol. 26, 101247 (2019).
Acknowledgements
The authors are grateful for funding from the European Research Council (ERC) (S.L.F., 788231; A.P.G., 757991; S.O., 679001; N.J.T., 742987), the Engineering and Physical Sciences Research Council (EPSRC) (S.L.F. and N.J.T., EP/S005226/1), the Biotechnology and Biological Sciences Research Council (BBSRC) (S.L.F. and N.J.T., BB/M027791/1, BB/M028836/1; A.P.G., BB/M027023/1), the Spanish Ministry of Economy and Competitiveness (MINECO) (S.O., PGC2018-102192-B-I00), Generalitat de Catalunya (S.O., SGR 2017 1707) and the University of Manchester (Presidential Fellowship to S.L.L.).
Author information
Authors and Affiliations
Contributions
Introduction (S.L.F., H.N. and K.S.R.); Experimentation (E.L.B., A.P.G., H.N., S.O., K.S.R., N.J.T., S.L.L., L.J.H. and W.F.); Results (M.A.H. and E.R.); Applications (S.P.F., M.A.H. and E.R.); Reproducibility and data deposition (W.F. and L.J.H.); Limitations and optimizations (W.F., L.J.H., M.A.H. and E.R.); Outlook (S.L.F.); overview of Primer (S.L.F.). All authors contributed equally to planning and revision of the manuscript as described. Please note that co-authors have been listed in alphabetical order.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Methods Primers thanks L. Betancor, A. Fryszkowska and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
BioCatNet: https://www.biocatnet.de/
Basic Local Alignment Search Tool : https://blast.ncbi.nlm.nih.gov/Blast.cgi
BRENDA: https://www.brenda-enzymes.org/
CAVER: http://www.caver.cz/
KEGG: https://www.genome.jp/kegg/
InterPro: https://www.ebi.ac.uk/interpro/
Pfam: https://pfam.xfam.org/
PrenDB: http://prendb.pharmazie.uni-marburg.de/prendb/home/
Protein Data Bank: https://www.rcsb.org/
RetroBioCat: https://retrobiocat.com/
Revenant: https://revenant.inf.pucp.edu.pe/
Rosetta: https://www.rosettacommons.org/
UniProt: https://www.uniprot.org/
Glossary
- Enzyme cascade
-
Within the biocatalysis community this term is used broadly for concurrent, multienzyme one-pot biocatalytic reactions as well as reactions in which components are added sequentially or process steps are telescoped.
- Metagenomic libraries
-
Genomic libraries constructed by the direct cloning of the large fragments of the environmental DNA into an appropriate vector, transformed into the host bacteria.
- C(sp 3)–H functionalization
-
A type of reaction in which a C–H bond, in which the carbon is sp3 hybridized, is cleaved and a new C–X bond is formed (where X is usually carbon, oxygen, nitrogen or a halide).
- Diels–Alderases
-
Enzymes that catalyse a [4 + 2] cycloaddition reaction between a conjugated diene and a substituted alkene forming a cyclohexene derivative.
- Rates of catalysis
-
The rates by which substrates are converted into products in catalytic reactions.
- Saturation mutagenesis
-
A method that allows the randomization of a target codon or set of codons in a gene.
- Iterative combinatorial active site testing
-
A method that allows the generation of DNA libraries where active site positions are randomized in pairs
- Error-prone PCR
-
A PCR (polymerase chain reaction) that is run under reaction conditions that introduce random mutations into the target DNA sequence.
- Gene shuffling
-
A method that allows for the generation of chimeric libraries of genes.
- DNA shuffling
-
A method that allows the recombination of beneficial mutations in a directed evolution experiment.
- High performance liquid chromatography
-
An analytical technique that allows for the rapid separation and quantification of compound mixtures using pressurized liquid solvent passed through chromatographic columns.
- Nonsense codon
-
A codon within the genetic code that does not encode an amino acid but is recognized as a stop codon in transcription and translation of DNA.
- Regioselectivity
-
The property that favours bond formation or breaking at a particular atom over all other possible atoms in a molecule.
- Enzyme operational stability
-
Retention of enzyme activity when the enzyme is in use.
- Evolvability
-
Capacity of an enzyme to acquire beneficial properties or functions through genetic modification.
- Design of experiments
-
A statistical approach to analyse the influence of various factors in a system to predict the optimal operating conditions.
- BLAST
-
(Basic Local Alignment Search Tool). A tool that compares nucleotide or protein sequences of interest (most commonly to sequences within a database), and finds regions of statistically significant similarity.
Rights and permissions
About this article
Cite this article
Bell, E.L., Finnigan, W., France, S.P. et al. Biocatalysis. Nat Rev Methods Primers 1, 46 (2021). https://doi.org/10.1038/s43586-021-00044-z
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43586-021-00044-z
This article is cited by
-
Bringing biocatalysis into teaching labs
Nature Chemistry (2026)
-
Advances in the sustainable biosynthesis of valuable terpenoid flavor compounds and precursors in micro-organisms
Biotechnology for Biofuels and Bioproducts (2025)
-
Cofactor-independent photo-enzymatic reductions with water mediated by reductive graphene quantum dots
Nature Communications (2025)
-
Engineered enzymes for enantioselective nucleophilic aromatic substitutions
Nature (2025)
-
Rational engineering of a thermostable α-oxoamine synthase biocatalyst expands the substrate scope and synthetic applicability
Communications Chemistry (2025)