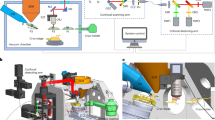
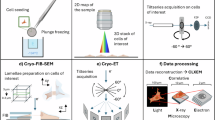
Abstract
The importance of cryogenic electron tomography (cryo-ET), particularly cryogenic cellular tomography, in uncovering the complexity of eukaryotic cell systems is only beginning to be fully recognized. As the only structural technique capable of generating detailed, three-dimensional visualizations of cellular architecture in situ at sub-nanometre resolution, cryo-ET offers unmatched insights into cell biology and disease mechanisms. Integrating cryo-ET with light microscopy techniques allows researchers to localize specific regions of interest, then directly correlate them with high-resolution structural data, enabling the investigation of cellular components and dynamic processes with a level of precision previously unattainable. Despite its transformative potential, mastering integrative light microscopy techniques and cryo-ET workflows remains highly challenging. These techniques require specialized equipment, extensive hands-on experience and a deep understanding of practical nuances, many of which are acquired only through practice. In this Primer, we highlight key considerations, tips and common pitfalls to help guide newcomers to these approaches as they navigate how to begin and what challenges to anticipate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 1 digital issues and online access to articles
$119.00 per year
only $119.00 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Chari, A. & Stark, H. Prospects and limitations of high-resolution single-particle cryo-electron microscopy. Annu. Rev. Biophys. 52, 391–411 (2023).
Bäuerlein, F. J. B. & Baumeister, W. Towards visual proteomics at high resolution. J. Mol. Biol. 433, 167187 (2021).
Beck, M. & Baumeister, W. Cryo-electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 26, 825–837 (2016).
Young, L. N. & Villa, E. Bringing structure to cell biology with cryo-electron tomography. Annu. Rev. Biophys. 52, 573–595 (2023).
Turk, M. & Baumeister, W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 594, 3243–3261 (2020).
Majumder, P. & Zhang, P. In situ cryo-electron microscopy and tomography of cellular and organismal samples. Curr. Opin. Struct. Biol. 93, 103076 (2025).
Nogales, E. & Mahamid, J. Bridging structural and cell biology with cryo-EM. Nature 628, 47–56 (2024).
Adrian, M., Dubochet, J., Lepault, J. & McDowall, A. W. Cryo-electron microscopy of viruses. Nature 308, 32–36 (1984).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988).
Sartori, A. et al. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160, 135–145 (2007).
Godman, G. C., Morgan, C., Breitenfeld, P. M. & Rose, H. M. A correlative study by electron and light microscopy of the development of type 5 adenovirus: II. Light microscopy. J. Exp. Med. 112, 383–402 (1960).
Morgan, C., Godman, G. C., Breitenfeld, P. M. & Rose, H. M. A correlative study by electron and light microscopy of the development of type 5 adenovirus: I. Electron microscopy. J. Exp. Med. 112, 373–382 (1960).
Dow, L. P. et al. Morphological control enables nanometer-scale dissection of cell-cell signaling complexes. Nat. Commun. 13, 7831 (2022).
Kim, J. Y. et al. Morphological comparison of primary neurons cryo-preserved under varied conditions. Microsc. Microanal. 29, 956–957 (2023).
Rigort, A. & Leis, A. in Cryo-electron Tomography A Journey from Sample Preparation to Data Mining (eds Hanein, D. & Volkmann, N.) 21–60 (Academic Press, 2025).
Toro-Nahuelpan, M. et al. Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies. Nat. Methods 17, 50–54 (2020).
Dobro, M. J., Melanson, L. A., Jensen, G. J. & McDowall, A. W. Plunge freezing for electron cryomicroscopy. Methods Enzymol. 481, 63–82 (2010).
Lam, V. & Villa, E. in cryoEM. Methods and Protocols Vol. 2215 (eds Gonen, T. & Nannenga, B.L.) 49–82 (Humana, 2021).
Marston, D. J. et al. High Rac1 activity is functionally translated into cytosolic structures with unique nanoscale cytoskeletal architecture. Proc. Natl Acad. Sci. USA 116, 1267–1272 (2019).
Fäßler, F., Zens, B., Hauschild, R. & Schur, F. K. M. 3D printed cell culture grid holders for improved cellular specimen preparation in cryo-electron microscopy. J. Struct. Biol. 212, 107633 (2020).
Aebi, U. & Pollard, T. D. A glow discharge unit to render electron microscope grids and other surfaces hydrophilic. J. Electron Microsc. Tech. 7, 29–33 (1987).
Isabell, T. C., Fischione, P. E., O’Keefe, C. & Guruz, M. U. & Dravid, V. P. Plasma cleaning and its applications for electron microscopy. Microsc. Microanal. 5, 126–135 (1999).
Gaietta, G. et al. Novel cryo-tomography workflow reveals nanometer-scale responses of epithelial cells to matrix stiffness and dimensionality. Mol. Biol. Cell 33, br28 (2022).
Engel, L. et al. Extracellular matrix micropatterning technology for whole cell cryogenic electron microscopy studies. J. Micromech. Microeng. 29, 115018 (2019).
Kai, F. et al. ECM dimensionality tunes actin tension to modulate endoplasmic reticulum function and spheroid phenotypes of mammary epithelial cells. EMBO J. 41, e109205 (2022).
Wu, G. H. et al. Multi-scale 3D cryo-correlative microscopy for vitrified cells. Structure 28, 1231–1237.e3 (2020).
Moravcová, J., Pinkas, M., Holbová, R. & Nováček, J. Preparation and cryo-FIB micromachining of saccharomyces cerevisiae for cryo-electron tomography. J. Vis. Exp. 2021, e62351 (2021).
Gaietta, G., Swift, M. F., Volkmann, N. & Hanein, D. Rapid tool for cell nanoarchitecture integrity assessment. J. Struct. Biol. 213, 107801 (2021).
Bailey, S. M. & Zasadzinski, J. A. N. Validation of convection-limited cooling of samples for freeze-fracture electron microscopy. J. Microsc. 163, 307–320 (1991).
Sartori, N., Richter, K. & Dubochet, J. Vitrification depth can be increased more than 10-fold by high-pressure freezing. J. Microsc. 172, 55–61 (1993).
Zhao, D. Y. et al. Autophagy preferentially degrades non-fibrillar polyQ aggregates. Mol. Cell 84, 1980–1994.e8 (2024).
Hsieh, C. E., Leith, A. D., Mannella, C. A., Frank, J. & Marko, M. Towards high-resolution three-dimensional imaging of native mammalian tissue: electron tomography of frozen-hydrated rat liver sections. J. Struct. Biol. 153, 1–13 (2006).
Kodera, C. et al. A systematic approach of vitrification by high pressure freezing. Methods Microsc. 1, 31–48 (2024).
Kelley, K. et al. Waffle method: a general and flexible approach for improving throughput in FIB-milling. Nat. Commun. 13, 1857 (2022).
Rubino, S. et al. A site-specific focused-ion-beam lift-out method for cryo transmission electron microscopy. J. Struct. Biol. 180, 572–576 (2012).
Schiøtz, O. H. et al. Serial lift-out: sampling the molecular anatomy of whole organisms. Nat. Methods 21, 1684–1692 (2024).
Hanein, D. & Volkmann, N. Correlative light-electron microscopy. Adv. Protein Chem. Struct. Biol. 82, 91–99 (2011).
Delorme-Walker, V. et al. Toxofilin upregulates the host cortical actin cytoskeleton dynamics, facilitating Toxoplasma invasion. J. Cell Sci. 125, 4333–4342 (2012).
Plitzko, J. M., Rigort, A. & Leis, A. Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes. Curr. Opin. Biotechnol. 20, 83–89 (2009).
Schwartz, C. L., Sarbash, V. I., Ataullakhanov, F. I., McIntosh, J. R. & Nicastro, D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J. Microsc. 227, 98–109 (2007).
Dahlberg, P. D. & Moerner, W. E. Cryogenic super-resolution fluorescence and electron microscopy correlated at the nanoscale. Annu. Rev. Phys. Chem. 72, 253–278 (2021).
Chang, Y. W. et al. Correlated cryogenic photoactivated localization microscopy and electron cryotomography. Nat. Methods 11, 737–739 (2014).
Perez, D. et al. Exploring transient states of PAmKate to enable improved cryogenic single-molecule imaging. J. Am. Chem. Soc. 146, 28707–28716 (2024).
Sartor, A. M., Dahlberg, P. D., Perez, D. & Moerner, W. E. Characterization of mApple as a red fluorescent protein for cryogenic single-molecule imaging with turn-off and turn-on active control mechanisms. J. Phys. Chem. B 127, 2690–2700 (2023).
Carter, S. D. et al. Distinguishing signal from autofluorescence in cryogenic correlated light and electron microscopy of mammalian cells. J. Struct. Biol. 201, 15–25 (2018).
Yu, P. S., Kim, C. U. & Lee, J. B. Cryogenic single-molecule fluorescence imaging. BMB Rep. 58, 2–7 (2025).
Szöllösi, J., Lockett, S. J., Balázs, M. & Waldman, F. M. Autofluorescence correction for fluorescence in situ hybridization. Cytometry 20, 356–361 (1995).
Mansfield, J. R., Gossage, K. W., Hoyt, C. C. & Levenson, R. M. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. J. Biomed. Opt. 10, 41207 (2005).
Fung, H. K. H. et al. Genetically encoded multimeric tags for subcellular protein localization in cryo-EM. Nat. Methods 20, 1900–1908 (2023).
Silvester, E. et al. DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell 184, 1110–1121.e16 (2021).
Sigmund, F. et al. Genetically encoded barcodes for correlative volume electron microscopy. Nat. Biotechnol. 41, 1734–1745 (2023).
Lim, S. D., Huang, Q. & Seibel, E. J. Evaluation of formalin fixation for tissue biopsies using shear wave laser speckle imaging system. IEEE J. Transl. Eng. Health Med. 7, 1500110 (2019).
Tuijtel, M. W., Koster, A. J., Jakobs, S., Faas, F. G. A. & Sharp, T. H. Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep. 9, 1369 (2019).
Last, M. G. F., Tuijtel, M. W., Voortman, L. M. & Sharp, T. H. Selecting optimal support grids for super-resolution cryogenic correlated light and electron microscopy. Sci. Rep. 13, 8270 (2023).
Waldchen, S., Lehmann, J., Klein, T., Van De Linde, S. & Sauer, M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 5, 15348 (2015).
Alghamdi, R. A., Exposito-Rodriguez, M., Mullineaux, P. M., Brooke, G. N. & Laissue, P. P. Assessing phototoxicity in a mammalian cell line: how low levels of blue light affect motility in PC3 cells. Front. Cell Dev. Biol. 9, 738786 (2021).
Henglein, A. Reactivity of free radicals at ambient and low temperatures. Ultramicroscopy 14, 195–200 (1984).
Dahlberg, P. D., Perez, D., Hecksel, C. W., Chiu, W. & Moerner, W. E. Metallic support films reduce optical heating in cryogenic correlative light and electron tomography. J. Struct. Biol. 214, 107901 (2022).
Hagen, W. J. H. Light ‘em up: efficient screening of gold foil grids in cryo-EM. Front. Mol. Biosci. 9, 912363 (2022).
Schorb, M. & Briggs, J. A. G. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy 143, 24–32 (2014).
Koning, R. I. et al. Correlative cryo-fluorescence light microscopy and cryo-electron tomography of Streptomyces. Methods Cell Biol. 124, 217–239 (2014).
Anderson, K. L., Page, C., Swift, M. F., Hanein, D. & Volkmann, N. Marker-free method for accurate alignment between correlated light, cryo-light, and electron cryo-microscopy data using sample support features. J. Struct. Biol. 201, 46–51 (2018).
Yang, J. E., Larson, M. R., Sibert, B. S., Shrum, S. & Wright, E. R. CorRelator: interactive software for real-time high precision cryo-correlative light and electron microscopy. J. Struct. Biol. 213, 107709 (2021).
McDowall, A. W. et al. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. J. Microsc. 131, 1–9 (1983).
Al-Amoudi, A., Norlen, L. P. O. & Dubochet, J. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J. Struct. Biol. 148, 131–135 (2004).
Li, S. et al. HOPE-SIM, a cryo-structured illumination fluorescence microscopy system for accurately targeted cryo-electron tomography. Commun. Biol. 6, 474 (2023).
de Beer, M. et al. Precise targeting for 3D cryo-correlative light and electron microscopy volume imaging of tissues using a FinderTOP. Commun. Biol. 6, 510 (2023).
de Beer, M. et al. Visualizing biological tissues: a multiscale workflow from live imaging to 3D Cryo-CLEM. Microsc. Microanal. 27, 11–12 (2021).
Kapteijn, R. et al. Endocytosis-like DNA uptake by cell wall-deficient bacteria. Nat. Commun. 13, 5524 (2022).
Faul, N. et al. Cryo-iCLEM: cryo correlative light and electron microscopy with immersion objectives. J. Struct. Biol. 217, 108179 (2025).
Li, S. et al. ELI trifocal microscope: a precise system to prepare target cryo-lamellae for in situ cryo-ET study. Nat. Methods 20, 276–283 (2023).
Li, W. et al. Integrated multimodality microscope for accurate and efficient target-guided cryo-lamellae preparation. Nat. Methods 20, 268–275 (2023).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Schellenberger, P. et al. High-precision correlative fluorescence and electron cryo microscopy using two independent alignment markers. Ultramicroscopy 143, 41–51 (2014).
Rodríguez de Francisco, B., Bezault, A., Xu, X. P., Hanein, D. & Volkmann, N. MEPSi: a tool for simulating tomograms of membrane-embedded proteins. J. Struct. Biol. 214, 107921 (2022).
Chua, E. Y. D. et al. Better, faster, cheaper: recent advances in cryo–electron microscopy. Annu. Rev. Biochem. 91, 1–32 (2022).
Kak, A. C. & Slaney, M. Principles of Computerized Tomographic Imaging (SIAM, 2001).
Radermacher, M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J. Electron Microsc. Tech. 9, 359–394 (1988).
De Rosier, D. J. & Klug, A. Reconstruction of three dimensional structures from electron micrographs. Nature 217, 130–134 (1968).
Gilbert, P. Iterative methods for the three-dimensional reconstruction of an object from projections. J. Theor. Biol. 36, 105–117 (1972).
Uddin, M. R., Dip, S. A., Jha, R. A., Zeng, X. & Xu, M. in Cryo-electron Tomography: A Journey from Sample Preparation to Data Mining (eds Hanein, D. & Volkmann, N.) 173–215 (Academic Press, 2025).
Förster, F. Subtomogram analysis: the sum of a tomogram’s particles reveals molecular structure in situ. J. Struct. Biol. X 6, 100063 (2022).
Tegunov, D., Xue, L., Dienemann, C., Cramer, P. & Mahamid, J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat. Methods 18, 186–193 (2021).
Xue, L. et al. Visualizing translation dynamics at atomic detail inside a bacterial cell. Nature 610, 205–211 (2022).
Volkmann, N. in Cryo-electron Tomography: A Journey from Sample Preparation to Data Mining (eds Hanein, D. & Volkmann, N.) 117–144 (Academic Press, 2025).
Anderson, K. L. et al. Nano-scale actin-network characterization of fibroblast cells lacking functional Arp2/3 complex. J. Struct. Biol. 197, 312–321 (2017).
Mageswaran, S. K., Yang, W. Y., Chakrabarty, Y., Oikonomou, C. M. & Jensen, G. J. A cryo-electron tomography workflow reveals protrusion-mediated shedding on injured plasma membrane. Sci. Adv. 7, eabc6345 (2021).
Kumar, A. et al. Local tension on talin in focal adhesions correlates with F-actin alignment at the nanometer scale. Biophys. J. 115, 1569–1579 (2018).
Suraneni, P. et al. A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol. Biol. Cell 26, 901–912 (2015).
Suraneni, P. et al. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197, 239–251 (2012).
Wolff, G. et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369, 1395–1398 (2020).
Liu, J. et al. The palisade layer of the poxvirus core is composed of flexible A10 trimers. Nat. Struct. Mol. Biol. 31, 1105–1113 (2024).
Mageswaran, S. K. et al. Nanoscale details of mitochondrial constriction revealed by cryoelectron tomography. Biophys. J. 122, 3768–3782 (2023).
Hou, Z., Nightingale, F., Zhu, Y., MacGregor-Chatwin, C. & Zhang, P. Structure of native chromatin fibres revealed by Cryo-ET in situ. Nat. Commun. 14, 6324 (2023).
Zhang, X. et al. Molecular mechanisms of stress-induced reactivation in mumps virus condensates. Cell 186, 1877–1894.e27 (2023).
Arnold, J. et al. Site-specific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys. J. 110, 860 (2016).
Liu, Y. et al. Cryo-electron tomography of NLRP3-activated ASC complexes reveals organelle co-localization. Nat. Commun. 14, 7246 (2023).
Matsui, A. et al. Cryo-electron tomographic investigation of native hippocampal glutamatergic synapses. eLife 13, RP98458 (2024).
Chiu, W., Schmid, M. F., Pintilie, G. D. & Lawson, C. L. Evolution of standardization and dissemination of cryo-EM structures and data jointly by the community, PDB, and EMDB. J. Biol. Chem. 296, 100560 (2021).
Bialy, N. et al. Harmonizing the Generation and Pre-publication Stewardship of FAIR Image Data. Preprint at https://doi.org/10.48550/arXiv.2401.13022 (2024).
Lawson, C. L. et al. EMDataBank unified data resource for 3DEM. Nucleic Acids Res. 44, D396–D403 (2016).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Iudin, A. et al. EMPIAR: the electron microscopy public image archive. Nucleic Acids Res. 51, D1503–D1511 (2023).
Hartley, M. et al. The bioimage archive - building a home for life-sciences microscopy data. J. Mol. Biol. 434, 167505 (2022).
Klumpe, S. et al. A modular platform for automated cryo-FIB workflows. eLife 10, e70506 (2021).
Cleeve, P. et al. OpenFIBSEM: a universal API for FIBSEM control. J. Struct. Biol. 215, 107967 (2023).
Zachs, T. et al. Fully automated, sequential focused ion beam milling for cryo-electron tomography. eLife 9, e52286 (2020).
Buckley, G. et al. Automated cryo-lamella preparation for high-throughput in-situ structural biology. J. Struct. Biol. 210, 107488 (2020).
Kuba, J. et al. Advanced cryo-tomography workflow developments – correlative microscopy, milling automation and cryo-lift-out. J. Microsc. 281, 112–124 (2021).
Tacke, S. et al. A streamlined workflow for automated cryo focused ion beam milling. J. Struct. Biol. 213, 107743 (2021).
Berger, C. et al. Plasma FIB milling for the determination of structures in situ. Nat. Commun. 14, 629 (2023).
Burnett, T. L. et al. Large volume serial section tomography by Xe plasma FIB dual beam microscopy. Ultramicroscopy 161, 119–129 (2016).
Weiner, A. et al. Vitrification of thick samples for soft X-ray cryo-tomography by high pressure freezing. J. Struct. Biol. 181, 77–81 (2013).
Zhu, H., Li, M., Li, M., Li, X. & Ou, G. Cryo-electron tomography elucidates annular intraluminal configurations in Caenorhabditis elegans microtubules. Biol. Cell 116, e2400064 (2024).
Parmenter, C. D. J., Fay, M. W., Hartfield, C. & Eltaher, H. M. Making the practically impossible “merely difficult”—cryogenic FIB lift-out for “damage free” soft matter imaging. Microsc. Res. Tech. 79, 298–303 (2016).
Woods, E. V. et al. A versatile and reproducible cryo-sample preparation methodology for atom probe studies. Microsc. Microanal. 29, 1992–2003 (2023).
Nguyen, H. T. D. et al. Serialized on-grid lift-in sectioning for tomography (SOLIST) enables a biopsy at the nanoscale. Nat. Methods 21, 1693–1701 (2024).
Xing, H. et al. Translation dynamics in human cells visualized at high resolution reveal cancer drug action. Science 381, 70–75 (2023).
Schiøtz, O. H., Klumpe, S., Plitzko, J. M. & Kaiser, C. J. O. Cryo-electron tomography: en route to the molecular anatomy of organisms and tissues. Biochem. Soc. Trans. 52, 2415–2425 (2024).
Berger, C. et al. Cryo-electron tomography on focused ion beam lamellae transforms structural cell biology. Nat. Methods 20, 499–511 (2023).
Fuest, M. et al. In situ microfluidic cryofixation for cryo focused ion beam milling and cryo electron tomography. Sci. Rep. 9, 19133 (2019).
Yoniles, J. et al. Time-resolved cryogenic electron tomography for the study of transient cellular processes. Mol. Biol. Cell 35, mr4 (2024).
Voss, J. M., Harder, O. F., Olshin, P. K., Drabbels, M. & Lorenz, U. J. Rapid melting and revitrification as an approach to microsecond time-resolved cryo-electron microscopy. Chem. Phys. Lett. 778, 138812 (2021).
Dumoux, M. et al. Cryo-plasma FIB/SEM volume imaging of biological specimens. eLife 12, e83623 (2023).
Klumpe, S. et al. In-cell structure and snapshots of copia retrotransposons in intact tissue by cryo-ET. Cell 188, 2094–2110.e18 (2025).
Eisenstein, F. et al. Parallel cryo electron tomography on in situ lamellae. Nat. Methods 20, 131–138 (2022).
Feldmüller, M. et al. Stepwise assembly and release of Tc toxins from Yersinia entomophaga. Nat. Microbiol. 9, 405–420 (2024).
Singh, D. et al. The molecular architecture of the nuclear basket. Cell 187, 5267–5281.e13 (2024).
Huang, Y. et al. Molecular architecture of coronavirus double-membrane vesicle pore complex. Nature 633, 224–231 (2024).
Eisenstein, F., Fukuda, Y. & Danev, R. Smart parallel automated cryo-electron tomography. Nat. Methods 21, 1612–1615 (2024).
Paul-Gilloteaux, P. et al. eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nat. Methods 14, 102–103 (2017).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Zheng, S. et al. AreTomo: an integrated software package for automated marker-free, motion-corrected cryo-electron tomographic alignment and reconstruction. J. Struct. Biol. X 6, 100068 (2022).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Agulleiro, J. I. & Fernandez, J. J. Fast tomographic reconstruction on multicore computers. Bioinformatics 27, 582–583 (2011).
Acknowledgements
The authors thank M. Lemos and A. Bezault (Institut Pasteur) for their contributions to parts of the data presented here. D.H. acknowledges funding from the US Army Research Office under contract W911NF-19-D-0001 for the Institute for Collaborative Biotechnologies award. C.S. thanks the ANR-FRANCE (French National Research Agency) for its financial support, project no. ANR-22-CPJ2-0040-01. The authors acknowledge funding from the Institut Pasteur and the CNRS (D.H. and N.V.), and the NanoImaging Core (NCF) at Institut Pasteur for the provision of the equipment (Vitrobot, and cryo-electron microscopes). NanoImaging Core was created with the help of a grant from the French Government’s Investissements d’Avenir program (EQUIPEX CACSICE - Centre d’analyse de systèmes complexes dans les environements complexes, ANR-11-EQPX-0008).
Author information
Authors and Affiliations
Contributions
Introduction (C.S., N.V. and D.H.); Experimentation (C.S., P.V.B., A.H., N.V., and D.H.); Results (C.S., P.V.B., A.H., N.V. and D.H.); Applications (C.S., P.V.B., A.H., N.V. and D.H.); Reproducibility and data deposition (C.S., N.V. and D.H.); Limitations and optimization (C.S., N.V. and D.H.); Outlook (C.S., N.V. and D.H.).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Methods Primers thanks Peter Dahlberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Bioimage Archive: https://www.ebi.ac.uk/bioimage-archive
EMDB: https://www.ebi.ac.uk/emdb
EMPIAR: https://www.ebi.ac.uk/empiar
Protein Data Bank (PDB): https://www.rcsb.org/
Supplementary information
Glossary
- Correlative light and electron microscopy
-
(CLEM). Workflows that combine or integrate light and electron microscopy to precisely localize and correlate dynamic cellular events with structural snapshots.
- Cryogenic focused ion beam milling
-
(Cryo-FIB milling). Process implemented to prepare lamellae at specific locations by ablating excess cellular material from the top and bottom of the sample.
- Devitrification
-
Transition of water from the vitreous (amorphous) state to crystalline ice or liquid water.
- Grids
-
Electron microscope supports made up of 3.05 mm-diameter circles, each consisting of a metal mesh coated with a thin film onto which samples are deposited for imaging.
- Holey carbon film
-
Film support made from carbon that contains perforations of defined size and spacing.
- Vitrification
-
Process of rapidly freezing water into its vitreous (amorphous) state, thereby preventing the formation of ice crystals.
- Waffle freezing
-
Cryogenic electron tomography freezing method that consists of using a high-pressure-freezing machine and allowing to freeze thicker samples for subsequent cryogenic electron tomography experiments.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sauvanet, C., Van Blerkom, P., Hatipoglu, A. et al. Correlative cryogenic light and electron tomography of eukaryotic cells. Nat Rev Methods Primers 5, 77 (2025). https://doi.org/10.1038/s43586-025-00447-2
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43586-025-00447-2