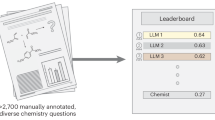
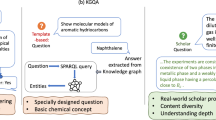
Abstract
Large language models (LLMs) offer opportunities for advancing chemical research, including planning, optimization, data analysis, automation and knowledge management. Deploying LLMs in active environments, where they interact with tools and data, can greatly enhance their capabilities. However, challenges remain in evaluating their performance and addressing ethical issues such as reproducibility, data privacy and bias. Here we discuss ongoing and potential integrations of LLMs in chemical research, highlighting existing challenges to guide the effective use of LLMs as active scientific partners.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
The Fourth Paradigm: Data-Intensive Scientific Discovery (Microsoft Research, 2009).
Meftahi, N. et al. Machine learning property prediction for organic photovoltaic devices. npj Comput. Mater. 6, 166 (2020).
Gupta, A., Chakraborty, S. & Ramakrishnan, R. Revving up 13C NMR shielding predictions across chemical space: benchmarks for atoms-in-molecules kernel machine learning with new data for 134 kilo molecules. Mach. Learn. Sci. Technol. 2, 035010 (2021).
Pinheiro, G. A. et al. Machine learning prediction of nine molecular properties based on the SMILES representation of the QM9 quantum-chemistry dataset. J. Phys. Chem. A 124, 9854–9866 (2020).
Guan, Y., Shree Sowndarya, S. V., Gallegos, L. C., St. John, P. C. & Paton, R. S. Real-time prediction of 1H and 13C chemical shifts with DFT accuracy using a 3D graph neural network. Chem. Sci. 12, 12012–12026 (2021).
Borlido, P. et al. Exchange–correlation functionals for band gaps of solids: benchmark, reparametrization and machine learning. npj Comput. Mater. 6, 96 (2020).
Ward, L. et al. matminer: an open source toolkit for materials data mining. Comput. Mater. Sci. 152, 60–69 (2018).
Deringer, V. L., Caro, M. A. & Csányi, G. Machine learning interatomic potentials as emerging tools for materials science. Adv. Mater. 31, 1902765 (2019).
Grambow, C. A., Pattanaik, L. & Green, W. H. Deep learning of activation energies. J. Phys. Chem. Lett. 11, 2992–2997 (2020).
Wen, M., Blau, S. M., Spotte-Smith, E. W. C., Dwaraknath, S. & Persson, K. A. BonDNet: a graph neural network for the prediction of bond dissociation energies for charged molecules. Chem. Sci. 12, 1858–1868 (2021).
Griffiths, R.-R. & Miguel Hernández-Lobato, J. Constrained Bayesian optimization for automatic chemical design using variational autoencoders. Chem. Sci. 11, 577–586 (2020).
Schweidtmann, A. M. et al. Machine learning meets continuous flow chemistry: automated optimization towards the Pareto front of multiple objectives. Chem. Eng. J. 352, 277–282 (2018).
Weston, L. et al. Named entity recognition and normalization applied to large-scale information extraction from the materials science literature. J. Chem. Inf. Model. 59, 3692–3702 (2019).
Huang, S. & Cole, J. M. BatteryDataExtractor: battery-aware text-mining software embedded with BERT models. Chem. Sci. 13, 11487–11495 (2022).
Musielewicz, J., Wang, X., Tian, T. & Ulissi, Z. FINETUNA: fine-tuning accelerated molecular simulations. Mach. Learn. Sci. Technol. 3, 03LT01 (2022).
Sultan, M. M. & Pande, V. S. Automated design of collective variables using supervised machine learning. J. Chem. Phys. 149, 094106 (2018).
Roch, L. M. et al. ChemOS: orchestrating autonomous experimentation. Sci. Robot. 3, eaat5559 (2018).
Boiko, D. A., MacKnight, R., Kline, B. & Gomes, G. Autonomous chemical research with large language models. Nature 624, 570–578 (2023).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 5998–6008 (2017).
OpenAI et al. GPT-4 technical report. Preprint at http://arxiv.org/abs/2303.08774 (2024).
Yin, S. et al. A survey on multimodal large language models. Natl Sci. Rev. 11, nwae403 (2024).
Gemini Team et al. Gemini: a family of highly capable multimodal models. Preprint at http://arxiv.org/abs/2312.11805 (2024).
Honda, S., Shi, S. & Ueda, H. R. SMILES Transformer: pre-trained molecular fingerprint for low data drug discovery. Preprint at https://doi.org/10.48550/arXiv.1911.04738 (2019).
MegaMolBART. GitHub https://github.com/NVIDIA/MegaMolBART (2022).
Sakano, K., Furui, K. & Ohue, M. NPGPT: natural product-like compound generation with GPT-based chemical language models. J. Supercomput. 81, 352 (2025).
Mazuz, E., Shtar, G., Shapira, B. & Rokach, L. Molecule generation using transformers and policy gradient reinforcement learning. Sci. Rep. 13, 8799 (2023).
Ouyang, L. et al. Training language models to follow instructions with human feedback. In Advances in Neural Information Processing Systems (eds Koyejo, S. et al.) Vol. 35, 27730–27744 (Curran Associates, Inc., 2022).
Rafailov, R. et al. Direct preference optimization: Your language model is secretly a reward model. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) Vol. 36, 53728–53741 (Curran Associates, Inc., 2023).
M. Bran, A. et al. Augmenting large language models with chemistry tools. Nat. Mach. Intell. 6, 525–535 (2024).
Ruan, Y. et al. An automatic end-to-end chemical synthesis development platform powered by large language models. Nat. Commun. 15, 10160 (2024).
McNaughton, A. D. et al. CACTUS: Chemistry Agent Connecting Tool-Usage to Science. ACS Omega 9, 46563–46573 (2024).
Hendrycks, D. et al. Measuring massive multitask language understanding. Preprint at http://arxiv.org/abs/2009.03300 (2021).
Templeton, A. et al. Scaling monosemanticity: extracting interpretable features from Claude 3 Sonnet. Transformer Circuits Thread https://transformer-circuits.pub/2024/scaling-monosemanticity/index.html (Anthropic, 2024).
Miret, S. & Krishnan, N. M. A. Are LLMs ready for real-world materials discovery? Preprint at https://doi.org/10.48550/arXiv.2402.05200 (2024).
Geva, M. et al. Did Aristotle use a laptop? A question answering benchmark with implicit reasoning strategies. Trans. Assoc. Comput. Linguist. 9, 346–361 (2021).
Gao, Y. et al. Retrieval-augmented generation for large language models: a survey. Preprint at http://arxiv.org/abs/2312.10997 (2024).
Lin, Y.-T. & Chen, Y.-N. LLM-Eval: unified multi-dimensional automatic evaluation for open-domain conversations with large language models. Preprint at http://arxiv.org/abs/2305.13711 (2023).
Guo, T. et al. What can large language models do in chemistry? A comprehensive benchmark on eight tasks. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) Vol. 36, 59662–59688 (Curran Associates, Inc., 2023).
Mirza, A. et al. Are large language models superhuman chemists? Preprint at http://arxiv.org/abs/2404.01475 (2024).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Sainz, O. et al. NLP evaluation in trouble: on the need to measure LLM data contamination for each benchmark. Preprint at http://arxiv.org/abs/2310.18018 (2023).
Sharma, M. et al. Towards understanding sycophancy in language models. Preprint at http://arxiv.org/abs/2310.13548 (2023).
Ranaldi, L. & Pucci, G. When large language models contradict humans? Large language models’ sycophantic behaviour. Preprint at http://arxiv.org/abs/2311.09410 (2024).
Schoenegger, P. & Park, P. S. Large language model prediction capabilities: evidence from a real-world forecasting tournament. Preprint at https://doi.org/10.48550/arXiv.2310.13014 (2023).
Liu, S., Chen, C., Qu, X., Tang, K. & Ong, Y.-S. Large language models as evolutionary optimizers. In 2024 IEEE Congress on Evolutionary Computation (CEC) 1–8 (IEEE, 2024).
Chiang, W.-L. et al. Chatbot Arena: an open platform for evaluating LLMs by human preference. Preprint at https://doi.org/10.48550/arXiv.2403.04132 (2024).
Mucci, T. & Stryker, C. What Is Artificial Superintelligence? https://www.ibm.com/think/topics/artificial-superintelligence (IBM, 2023).
Brockman, G. et al. OpenAI Gym. Preprint at https://doi.org/10.48550/arXiv.1606.01540 (2016).
Wang, J. et al. GTA: a benchmark for general tool agents. Preprint at https://doi.org/10.48550/arXiv.2407.08713 (2024).
Qin, Y. et al. ToolLLM: facilitating large language models to master 16000+ real-world APIs. Preprint at https://doi.org/10.48550/arXiv.2307.16789 (2023).
Patil, S. G., Zhang, T., Wang, X. & Gonzalez, J. E. Gorilla: Large langage model connected with massive APIs. In Advances in Neural Information Processing Systems (eds Globerson, A. et al.) Vol. 37, 126544–126565 (Curran Associates, Inc., 2024).
Valmeekam, K., Marquez, M., Olmo, A., Sreedharan, S. & Kambhampati, S. PlanBench: An extensible benchmark for evaluating large language models on planning and reasoning about change. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) Vol. 36, 38975–38987 (Curran Associates, Inc., 2023).
Valmeekam, K., Marquez, M., Sreedharan, S. & Kambhampati, S. On the planning abilities of large language models—a critical investigation. Adv. Neural Inf. Process. Syst. 36, 75993–76005 (2023).
Skarlinski, M. D. et al. Language agents achieve superhuman synthesis of scientific knowledge. Preprint at https://doi.org/10.48550/arXiv.2409.13740 (2024).
Si, C., Yang, D. & Hashimoto, T. Can LLMs generate novel research ideas? A large-scale human study with 100+ NLP researchers. Preprint at https://doi.org/10.48550/arXiv.2409.04109 (2024).
HasAnyone https://platform.futurehouse.org/ (FutureHouse, 2024).
Zhou, Y., Liu, H., Srivastava, T., Mei, H. & Tan, C. Hypothesis generation with large language models. Preprint at https://doi.org/10.48550/arXiv.2404.04326 (2024).
Wellawatte, G. P. & Schwaller, P. Extracting human interpretable structure–property relationships in chemistry using XAI and large language models. Preprint at https://doi.org/10.48550/arXiv.2311.04047 (2023).
Learning to reason with LLMs. OpenAI https://openai.com/index/learning-to-reason-with-llms/ (2024)
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. In Advances in Neural Information Processing Systems (eds Koyejo, S. et al.) Vol. 35, 24824–24837 (Curran Associates, Inc., 2022).
Muralidharan, S. et al. Compact language models via pruning and knowledge distillation. In Advances in Neural Information Processing Systems (eds Globerson, A. et al.) Vol. 37, 41076–41102 (Curran Associates, Inc., 2024).
Sreenivas, S. T. et al. LLM pruning and distillation in practice: the Minitron approach. Preprint at https://doi.org/10.48550/arXiv.2408.11796 (2024).
Rai, D., Zhou, Y., Feng, S., Saparov, A. & Yao, Z. A practical review of mechanistic interpretability for transformer-based language models. Preprint at https://doi.org/10.48550/arXiv.2407.02646 (2024).
Zhou, Z., Li, X. & Zare, R. N. Optimizing chemical reactions with deep reinforcement learning. ACS Cent. Sci. 3, 1337–1344 (2017).
Bowden, G. D., Pichler, B. J. & Maurer, A. A design of experiments (DoE) approach accelerates the optimization of copper-mediated 18F-fluorination reactions of arylstannanes. Sci. Rep. 9, 11370 (2019).
Cao, B. et al. How to optimize materials and devices via design of experiments and machine learning: demonstration using organic photovoltaics. ACS Nano 12, 7434–7444 (2018).
Reis, M. et al. Machine-learning-guided discovery of 19F MRI agents enabled by automated copolymer synthesis. J. Am. Chem. Soc. 143, 17677–17689 (2021).
Mahjour, B., Hoffstadt, J. & Cernak, T. Designing chemical reaction arrays using Phactor and ChatGPT. Org. Process Res. Dev. 27, 1510–1516 (2023).
Přichystal, J., Schug, K. A., Lemr, K., Novák, J. & Havlíček, V. Structural analysis of natural products. Anal. Chem. 88, 10338–10346 (2016).
Nature submission guidelines. Nature Medicine https://www.nature.com/nm/submission-guidelines/aip-and-formatting (2025)
Yang, E. et al. Model merging in LLMs, MLLMs, and beyond: methods, theories, applications and opportunities. Preprint at https://doi.org/10.48550/arXiv.2408.07666 (2024).
Christensen, M. et al. Automation isn’t automatic. Chem. Sci. 12, 15473–15490 (2021).
Arnold, C. Cloud labs: where robots do the research. Nature 606, 612–613 (2022).
Liu, J., Xia, C. S., Wang, Y. & ZHANG, L. Is your code generated by ChatGPT really correct? Rigorous evaluation of large language models for code generation. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) vol. 36, 21558–21572 (Curran Associates, Inc., 2023).
O’Donoghue, O. et al. BioPlanner: automatic evaluation of LLMs on protocol planning in biology. Preprint at http://arxiv.org/abs/2310.10632 (2023).
Touvron, H. et al. LLaMA: open and efficient foundation language models. Preprint at https://doi.org/10.48550/arXiv.2302.13971 (2023).
Touvron, H. et al. Llama 2: open foundation and fine-tuned chat models. Preprint at https://doi.org/10.48550/arXiv.2307.09288 (2023).
Taylor, R. et al. Galactica: a large language model for science. Preprint at https://doi.org/10.48550/arXiv.2211.09085 (2022).
Zheng, Z., Zhang, O., Borgs, C., Chayes, J. T. & Yaghi, O. M. ChatGPT Chemistry Assistant for text mining and prediction of MOF synthesis. J. Am. Chem. Soc. 145, 18048–18062 (2023).
Perplexity AI. www.perplexity.ai (2022)
Jablonka, K. M., Schwaller, P., Ortega-Guerrero, A. & Smit, B. Leveraging large language models for predictive chemistry. Nat. Mach. Intell. 6, 161–169 (2024).
Dunn, A., Wang, Q., Ganose, A., Dopp, D. & Jain, A. Benchmarking materials property prediction methods: the Matbench test set and Automatminer reference algorithm. npj Comput. Mater. 6, 138 (2020).
Mateiu, P. & Groza, A. Ontology engineering with Large Language Models. Preprint at https://doi.org/10.48550/arXiv.2307.16699 (2023).
Babaei Giglou, H., D’Souza, J. & Auer, S. LLMs4OL: large language models for ontology learning. In The Semantic Web—ISWC 2023 (eds. Payne, T. R. et al.) 408–427 (Springer Nature, 2023).
Ciatto, G., Agiollo, A., Magnini, M. & Omicini, A. Large language models as oracles for instantiating ontologies with domain-specific knowledge. Knowl.-Based Syst. 310, 112940 (2025).
Ye, Y. et al. Construction and application of materials knowledge graph in multidisciplinary materials science via large language model. In Advances in Neural Information Processing Systems (eds Globerson, A. et al.) Vol. 37, 56878–56897 (Curran Associates, Inc., 2024).
Pascazio, L. et al. Chemical species ontology for data integration and knowledge discovery. J. Chem. Inf. Model. 63, 6569–6586 (2023).
Gontier, N., Rodriguez, P., Laradji, I., Vazquez, D. & Pal, C. Language decision transformers with exponential tilt for interactive text environments. Preprint at http://arxiv.org/abs/2302.05507 (2023).
Wu, Y.-H., Wang, X. & Hamaya, M. Elastic decision transformer. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) Vol. 36, 18532–18550 (Curran Associates, Inc., 2023).
Xi, Z. et al. The rise and potential of large language model based agents: a survey. Sci. China Inf. Sci. 68, 121101 (2025).
DeepSeek-AI et al. DeepSeek-R1: incentivizing reasoning capability in LLMs via reinforcement learning. Preprint at https://doi.org/10.48550/arXiv.2501.12948 (2025).
Wang, X. et al. Executable code actions elicit better LLM agents. Preprint at http://arxiv.org/abs/2402.01030 (2024).
Zhang, B. et al. Benchmarking the text-to-SQL capability of large language models: a comprehensive evaluation. Preprint at http://arxiv.org/abs/2403.02951 (2024).
Cheng, G. et al. Empowering large language models on robotic manipulation with affordance prompting. Preprint at http://arxiv.org/abs/2404.11027 (2024).
Reaxys. https://reaxys.com (Elsevier, 2009).
SciFinder. https://scifinder-n.cas.org (CAS, 1995).
Swain, M. C. & Cole, J. M. ChemDataExtractor: a toolkit for automated extraction of chemical information from the scientific literature. J. Chem. Inf. Model. 56, 1894–1904 (2016).
Coley, C. W., Rogers, L., Green, W. H. & Jensen, K. F. Computer-assisted retrosynthesis based on molecular similarity. ACS Cent. Sci. 3, 1237–1245 (2017).
CVE-2023-36258 Detail. National Vulnerability Database (NIST, accessed 11 June 2024); https://nvd.nist.gov/vuln/detail/CVE-2023-36258
Ruan, Y. et al. Accelerated end-to-end chemical synthesis development with large language models. Preprint at https://doi.org/10.26434/chemrxiv-2024-6wmg4 (2024).
Tom, G. et al. Self-driving laboratories for chemistry and materials science. Chem. Rev. 124, 9633–9732 (2024).
Software Business: 14th International Conference, ICSOB 2023, Lahti, Finland, November 27–29, 2023, Proc. (Springer Nature, 2024).
Favato, D., Ishitani, D., Oliveira, J. & Figueiredo, E. Linus’s law: more eyes fewer flaws in open source projects. In Proc. XVIII Brazilian Symposium on Software Quality 69–78 (ACM, 2019).
Yao, S. et al. ReAct: synergizing reasoning and acting in language models. Preprint at https://doi.org/10.48550/arXiv.2210.03629 (2023).
Huang, Q., Vora, J., Liang, P. & Leskovec, J. MLAgentBench: evaluating language agents on machine learning experimentation. Preprint at https://doi.org/10.48550/arXiv.2310.03302 (2023).
Liu, X. et al. AgentBench: evaluating LLMs as agents. Preprint at https://doi.org/10.48550/arXiv.2308.03688 (2023).
Hasselgren, C. & Oprea, T. I. Artificial intelligence for drug discovery: are we there yet? Annu. Rev. Pharmacol. Toxicol. 64, 527–550 (2024).
Bordukova, M., Makarov, N., Rodriguez-Esteban, R., Schmich, F. & Menden, M. P. Generative artificial intelligence empowers digital twins in drug discovery and clinical trials. Expert Opin. Drug Discov. 19, 33–42 (2024).
AI’s potential to accelerate drug discovery needs a reality check. Nature 622, 217 (2023).
Zhang, Y. et al. Siren’s song in the AI ocean: a survey on hallucination in large language models. Preprint at https://doi.org/10.48550/arXiv.2309.01219 (2023).
Li, J., Cheng, X., Zhao, X., Nie, J.-Y. & Wen, J.-R. HaluEval: a large-scale hallucination evaluation benchmark for large language models. In Proc. 2023 Conference on Empirical Methods in Natural Language Processing 6449–6464 (Association for Computational Linguistics, 2023).
Dhuliawala, S. et al. Chain-of-Verification reduces hallucination in large language models. Preprint at https://doi.org/10.48550/arXiv.2309.11495 (2023).
Tonmoy, S. M. T. I. et al. A comprehensive survey of hallucination mitigation techniques in large language models. Preprint at https://doi.org/10.48550/arXIiv.2401.01313 (2024).
Liu, H. et al. A survey on hallucination in large vision-language models. Preprint at https://doi.org/10.48550/arXiv.2402.00253 (2024).
Guan, X. et al. Mitigating large language model hallucinations via autonomous knowledge graph-based retrofitting. In Proc. 38th AAAI Conference on Artificial Intelligence (eds Wooldridge, M. et al.) 18126–18134 (AAAI, 2024).
Xu, Z., Jain, S. & Kankanhalli, M. Hallucination is inevitable: an innate limitation of large language models. Preprint at https://doi.org/10.48550/arXiv.2401.11817 (2024).
Li, J. et al. The dawn after the dark: an empirical study on factuality hallucination in large language models. Preprint at https://doi.org/10.48550/arXiv.2401.03205 (2024).
Luo, J., Xiao, C. & Ma, F. Zero-resource hallucination prevention for large language models. Preprint at https://doi.org/10.48550/arXiv.2309.02654 (2023).
Zhang, D. et al. ChemLLM: a chemical large language model. Preprint at https://doi.org/10.48550/arXiv.2402.06852 (2024).
Yasunaga, M., Ren, H., Bosselut, A., Liang, P. & Leskovec, J. QA-GNN: reasoning with language models and knowledge graphs for question answering. Preprint at https://doi.org/10.48550/arXiv.2104.06378 (2021).
Lu, L. et al. Physics-informed neural networks with hard constraints for inverse design. SIAM J. Sci. Comput. 43, B1105–B1132 (2021).
Han, S. et al. LLM multi-agent systems: challenges and open problems. Preprint at http://arxiv.org/abs/2402.03578 (2024).
Darvish, K. et al. ORGANA: A robotic assistant for automated chemistry experimentation and characterization. Matter 8, 101897 (2025).
Formica, M. et al. Catalytic enantioselective nucleophilic desymmetrization of phosphonate esters. Nat. Chem. 15, 714–721 (2023).
Ramos, M. C., Collison, C. J. & White, A. D. A review of large language models and autonomous agents in chemistry. Chem. Sci. 16, 2514–2572 (2025).
Bran, A. M. & Schwaller, P. in Drug Development Supported by Informatics (eds Satoh, H. et al.) 143–163 (Springer Nature, 2024).
Pei, Q. et al. BioT5: enriching cross-modal integration in biology with chemical knowledge and natural language associations. Preprint at https://doi.org/10.48550/arXiv.2310.07276 (2024).
Fang, J. et al. MolTC: towards molecular relational modeling in language models. Preprint at https://doi.org/10.48550/arXiv.2402.03781 (2024).
Schuhmann, C. et al. LAION-5B: An open large-scale dataset for training next generation image-text models. In Advances in Neural Information Processing Systems (eds. Koyejo, S. et al.) Vol. 35, 25278–25294 (Curran Associates, Inc., 2022).
Huang, J., Shao, H. & Chang, K. C.-C. Are large pre-trained language models leaking your personal information? Preprint at https://doi.org/10.48550/arXiv.2205.12628 (2022).
Wahle, J. P., Ruas, T., Kirstein, F. & Gipp, B. How large language models are transforming machine-paraphrase plagiarism. In Proc. 2022 Conference on Empirical Methods in Natural Language Processing 952–963 (Association for Computational Linguistics, 2022).
Karamolegkou, A., Li, J., Zhou, L. & Søgaard, A. Copyright violations and large language models. Preprint at https://doi.org/10.48550/arXiv.2310.13771 (2023).
McDonald, J. et al. Great power, great responsibility: recommendations for reducing energy for training language models. In Findings of the Association for Computational Linguistics: NAACL 2022 1962–1970 (Association for Computational Linguistics, 2022).
Samsi, S. et al. From words to watts: benchmarking the energy costs of large language model inference. In 2023 IEEE High Performance Extreme Computing Conference (HPEC) 1–9 (IEEE, 2023).
Patterson, D. et al. Carbon emissions and large neural network training. Preprint at https://doi.org/10.48550/arXiv.2104.10350 (2021).
Omiye, J. A., Lester, J. C., Spichak, S., Rotemberg, V. & Daneshjou, R. Large language models propagate race-based medicine. npj Digit. Med. 6, 195 (2023).
The United States Artificial Intelligence Safety Institute: Vision, Mission, and Strategic Goals (NIST, 2024).
Canadian Artificial Intelligence Safety Institute (Government of Canada, accessed 30 January 2025); https://ised-isde.canada.ca/site/ised/en/canadian-artificial-intelligence-safety-institute
AISI Research (AI Security Institute, accessed 30 January 2025); https://www.aisi.gov.uk/category/research
Lee, S. & Manthiram, A. Can cobalt be eliminated from lithium-ion batteries? ACS Energy Lett. 7, 3058–3063 (2022).
Chung, C. et al. Decarbonizing the chemical industry: a systematic review of sociotechnical systems, technological innovations, and policy options. Energy Res. Soc. Sci. 96, 102955 (2023).
Xia, R., Overa, S. & Jiao, F. Emerging electrochemical processes to decarbonize the chemical industry. JACS Au 2, 1054–1070 (2022).
Amodei, D. Machines of Loving Grace https://darioamodei.com/machines-of-loving-grace (2024).
McNally, A., Prier, C. K. & MacMillan, D. W. C. Discovery of an α-amino C–H arylation reaction using the strategy of accelerated serendipity. Science 334, 1114–1117 (2011).
Ban, T. A. The role of serendipity in drug discovery. Dialogues Clin. Neurosci. 8, 335–344 (2006).
Acknowledgements
We thank J. Kitchin and E. Spotte-Smith (both at CMU Chemical Engineering) for their vital feedback while writing the manuscript. We gratefully acknowledge the financial support by the National Science Foundation Center for Computer-Assisted Synthesis (NSF C-CAS, grant no. 2202693). R.M. thanks CMU and its benefactors for the 2024–25 Tata Consulting Services Presidential Fellowship. J.E.R. thanks NSF and C-CAS for the 2024-25 DARE Fellowship. L.C.G. thanks the CMU Mellon College of Sciences for the Automated Sciences Postdoctoral Fellowship. Any opinions, findings and conclusions or recommendations expressed in this perspective are those of the author(s) and do not necessarily reflect the views of any of the funding sources.
Author information
Authors and Affiliations
Contributions
All authors participated in the writing of this manuscript.
Corresponding author
Ethics declarations
Competing interests
G.G. and R.M. are co-founders of evals, a consultancy firm for scientific evaluations of frontier AI models. The other authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Berend Smit and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
MacKnight, R., Boiko, D.A., Regio, J.E. et al. Rethinking chemical research in the age of large language models. Nat Comput Sci 5, 715–726 (2025). https://doi.org/10.1038/s43588-025-00811-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-025-00811-y