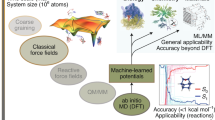
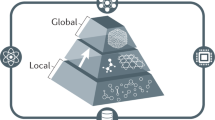
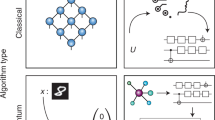
Abstract
As quantum mechanics marks its centennial in 2025, machine learning interatomic potentials have emerged as transformative tools in molecular modeling, bridging quantum mechanical accuracy with classical efficiency. Here we examine their development through four defining challenges—achieving chemical accuracy, maintaining computational efficiency, ensuring interpretability and reaching universal generalizability. We highlight architectural innovations, physics-informed approaches, and foundation models trained on extensive data. Together, these developments chart a path toward predictive, transferable and physically grounded machine learning frameworks for next-generation computational chemistry.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
About the International Year of Quantum Science and Technology https://www.unesco.org/en/quantum-science-technology/about (UNESCO, 2025).
Quantum mechanics 100 years on: an unfinished revolution. Nature 637, 251–252 (2025).
Feynman, R. P. Simulating physics with computers. Int. J. Theor. Phys. 21, 467–488 (1982).
Trabesinger, A. Quantum simulation. Nat. Phys. 8, 263 (2012).
Pople, J. A. Nobel lecture: quantum chemical models. Rev. Mod. Phys. 71, 1267–1274 (1999).
MacFarlane, A. G. J., Dowling, J. P. & Milburn, G. J. Quantum technology: the second quantum revolution. Philos. Trans. R. Soc. Lond. Ser. A 361, 1655–1674 (2003).
Preskill, J. Quantum computing in the NISQ era and beyond. Quantum 2, 79 (2018).
Venkatasubramanian, V. Celebrating the birth centenary of quantum mechanics: a historical perspective. Ind. Eng. Chem. Res. 64, 9443–9456 (2025).
Schrödinger, E. An undulatory theory of the mechanics of atoms and molecules. Phys. Rev. 28, 1049–1070 (1926).
Born, M. & Oppenheimer, R. Zur Quantentheorie der Molekeln. Ann. Phys. 389, 457–484 (1927).
Hohenberg, P. & Kohn, W. Inhomogeneous electron gas. Phys. Rev. 136, B864–B871 (1964).
Kohn, W. & Sham, L. J. Self-consistent equations including exchange and correlation effects. Phys. Rev. 140, A1133–A1138 (1965).
Jensen, F. Introduction to Computational Chemistry (Wiley, 2017).
Mardirossian, N. & Head-Gordon, M. Thirty years of density functional theory in computational chemistry: an overview and extensive assessment of 200 density functionals. Mol. Phys. 115, 2315–2372 (2017).
Keith, J. A. et al. Combining machine learning and computational chemistry for predictive insights into chemical systems. Chem. Rev. 121, 9816–9872 (2021).
Jacobs, R. et al. A practical guide to machine learning interatomic potentials – status and future. Curr. Opin. Solid State Mater. Sci. 35, 101214 (2025).
Butler, K. T., Davies, D. W., Cartwright, H., Isayev, O. & Walsh, A. Machine learning for molecular and materials science. Nature 559, 547–555 (2018).
Yang, X., Wang, Y., Byrne, R., Schneider, G. & Yang, S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem. Rev. 119, 10520–10594 (2019).
Coley, C. W., Green, W. H. & Jensen, K. F. Machine learning in computer-aided synthesis planning. Acc. Chem. Res. 51, 1281–1289 (2018).
Yang, K. et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59, 3370–3388 (2019).
Kirkpatrick, J. et al. Pushing the frontiers of density functionals by solving the fractional electron problem. Science 374, 1385–1389 (2021).
Hermann, J., Schätzle, Z. & Noé, F. Deep-neural-network solution of the electronic Schrödinger equation. Nat. Chem. 12, 891–897 (2020).
Pfau, D., Spencer, J. S., Matthews, A. G. D. G. & Foulkes, W. M. C. Ab initio solution of the many-electron Schrödinger equation with deep neural networks. Phys. Rev. Res. 2, 033429 (2020).
Hopfield, J. J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl Acad. Sci. USA 79, 2554–2558 (1982).
Ackley, D. H., Hinton, G. E. & Sejnowski, T. J. A learning algorithm for Boltzmann machines. Cogn. Sci. 9, 147–169 (1985).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Rumelhart, D. E., Hinton, G. E. & Williams, R. J. Learning representations by back-propagating errors. Nature 323, 533–536 (1986).
Otto, M. & Hörchner, U. in Software Development in Chemistry 4 (ed. Gasteiger, J.) 377–384 (Springer, 1990); https://doi.org/10.1007/978-3-642-75430-2_39.
Curry, B. & Rumelhart, D. E. MSnet: a neural network which classifies mass spectra. Tetrahedron Comput. Methodol. 3, 213–237 (1990).
Qian, N. & Sejnowski, T. J. Predicting the secondary structure of globular proteins using neural network models. J. Mol. Biol. 202, 865–884 (1988).
Holley, L. H. & Karplus, M. Protein secondary structure prediction with a neural network. Proc. Natl Acad. Sci. USA 86, 152–156 (1989).
Kireev, D. B. ChemNet: a novel neural network based method for graph/property mapping. J. Chem. Inf. Comput. Sci. 35, 175–180 (1995).
Wu, Z. et al. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 32, 4–24 (2021).
Corso, G., Stark, H., Jegelka, S., Jaakkola, T. & Barzilay, R. Graph neural networks. Nat. Rev. Methods Prim. 4, 17 (2024).
Blank, T. B., Brown, S. D., Calhoun, A. W. & Doren, D. J. Neural network models of potential energy surfaces. J. Chem. Phys. 103, 4129–4137 (1995).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 146401 (2007).
Deringer, V. L., Caro, M. A. & Csányi, G. Machine learning interatomic potentials as emerging tools for materials science. Adv. Mater. 31, 1902765 (2019).
Zhang, Y.-W. et al. Roadmap for the development of machine learning-based interatomic potentials. Model. Simul. Mater. Sci. Eng. 33, 023301 (2025).
Peterson, K. A., Feller, D. & Dixon, D. A. Chemical accuracy in ab initio thermochemistry and spectroscopy: current strategies and future challenges. Theor. Chem. Acc. 131, 1079 (2012).
Martin, J. M. L. & Santra, G. Empirical double-hybrid density functional theory: a ‘third way’ in between WFT and DFT. Isr. J. Chem. 60, 787–804 (2020).
Raghavachari, K., Trucks, G. W., Pople, J. A. & Head-Gordon, M. A fifth-order perturbation comparison of electron correlation theories. Chem. Phys. Lett. 157, 479–483 (1989).
Feller, D., Peterson, K. A. & Grant Hill, J. On the effectiveness of CCSD(T) complete basis set extrapolations for atomization energies. J. Chem. Phys. 135, 044102 (2011).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1, a data set of 20 million calculated off-equilibrium conformations for organic molecules. Sci. Data 4, 170193 (2017).
Devereux, C. et al. Extending the applicability of the ANI deep learning molecular potential to sulfur and halogens. J. Chem. Theory Comput. 16, 4192–4202 (2020).
Bronstein, M. M., Bruna, J., Cohen, T. & Veličković, P. Geometric deep learning: grids, groups, graphs, geodesics and gauges. Preprint at https://arxiv.org/abs/2104.13478 (2021).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In Proceedings of the 34th International Conference on Machine Learning, Vol. 70, 1263–1272 (PMLR, 2017).
Schütt, K. T., Sauceda, H. E., Kindermans, P.-J., Tkatchenko, A. & Müller, K.-R. SchNet – a deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018).
Gasteiger, J., Groß, J. & Günnemann, S. Directional message passing for molecular graphs. Preprint at https://arxiv.org/abs/2003.03123 (2022).
Gasteiger, J., Becker, F. & Günnemann, S. GemNet: universal directional graph neural networks for molecules. In Advances in Neural Information Processing Systems, Vol. 34, 6790–6802 (Curran Associates, Inc., 2021).
Chmiela, S., Sauceda, H. E., Poltavsky, I., Müller, K.-R. & Tkatchenko, A. sGDML: constructing accurate and data efficient molecular force fields using machine learning. Comput. Phys. Commun. 240, 38–45 (2019).
Lubbers, N., Smith, J. S. & Barros, K. Hierarchical modeling of molecular energies using a deep neural network. J. Chem. Phys. 148, 241715 (2018).
Kondor, R., Son, H. T., Pan, H., Anderson, B. & Trivedi, S. Covariant compositional networks for learning graphs. Preprint at https://arxiv.org/abs/1801.02144 (2018).
Thomas, N. et al. Tensor field networks: rotation- and translation-equivariant neural networks for 3D point clouds. Preprint at https://arxiv.org/abs/1802.08219 (2018).
Geiger, M. & Smidt, T. E3nn: euclidean neural networks. Preprint at https://arxiv.org/abs/2207.09453 (2022).
Haghighatlari, M. et al. NewtonNet: a Newtonian message passing network for deep learning of interatomic potentials and forces. Digit. Discov. 1, 333–343 (2022).
Batzner, S. et al. E(3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. Nat. Commun. 13, 2453 (2022).
Batatia, I., Kovács, D. P., Simm, G. N. C., Ortner, C. & Csányi, G. MACE: higher order equivariant message passing neural networks for fast and accurate force fields. In Advances in Neural Information Processing Systems, Vol. 35, 11423–11436 (Curran Associates, Inc., 2022).
Schütt, K. T., Unke, O. T. & Gastegger, M. Equivariant message passing for the prediction of tensorial properties and molecular spectra. In Proceedings of the 38th International Conference on Machine Learning, Vol. 139, 9377–9388 (PMLR, 2021).
Musaelian, A. et al. Learning local equivariant representations for large-scale atomistic dynamics. Nat. Commun. 14, 579 (2023).
Kovács, D. P. et al. MACE-OFF: short-range transferable machine learning force fields for organic molecules. J. Am. Chem. Soc. 147, 17598–1761 (2025).
Fu, X. et al. Learning smooth and expressive interatomic potentials for physical property prediction. In Proceedings of the 42nd International Conference on Machine Learning, Vol. 267, 17875–17893 (PMLR, 2025).
Wood, B. M. et al. UMA: a family of universal models for atoms. Preprint at https://arxiv.org/abs/2506.23971 (2025).
Jacobs, R. A., Jordan, M. I. & Barto, A. G. Task decomposition through competition in a modular connectionist architecture: the what and where vision tasks. Cogn. Sci. 15, 219–250 (1991).
Goerigk, L. et al. A look at the density functional theory zoo with the advanced GMTKN55 database for general main group thermochemistry, kinetics and noncovalent interactions. Phys. Chem. Chem. Phys. 19, 32184–32215 (2017).
Gould, T. & Dale, S. G. Poisoning density functional theory with benchmark sets of difficult systems. Phys. Chem. Chem. Phys. 24, 6398–6403 (2022).
Burke, K. Perspective on density functional theory. J. Chem. Phys. 136, 150901 (2012).
Cohen, A. J., Mori-Sánchez, P. & Yang, W. Challenges for density functional theory. Chem. Rev. 112, 289–320 (2012).
Wang, T. Y., Neville, S. P. & Schuurman, M. S. Machine learning seams of conical intersection: a characteristic polynomial approach. J. Phys. Chem. Lett. 14, 7780–7786 (2023).
Smith, J. S. et al. The ANI-1ccx and ANI-1x data sets, coupled-cluster and density functional theory properties for molecules. Sci. Data 7, 134 (2020).
Yang, Y., Eldred, M. S., Zádor, J. & Najm, H. N. Multifidelity neural network formulations for prediction of reactive molecular potential energy surfaces. J. Chem. Inf. Model. 63, 2281–2295 (2023).
Zheng, P., Zubatyuk, R., Wu, W., Isayev, O. & Dral, P. O. Artificial intelligence-enhanced quantum chemical method with broad applicability. Nat. Commun. 12, 7022 (2021).
Chen, Y. & Dral, P. O. AIQM2: organic reaction simulations beyond DFT. Chem. Sci. 16, 15901–15912 (2025).
Thaler, S., Gabellini, C., Shenoy, N. & Tossou, P. Implicit delta learning of high fidelity neural network potentials. Preprint at https://arxiv.org/abs/2412.06064 (2024).
Smith, J. S. et al. Approaching coupled cluster accuracy with a general-purpose neural network potential through transfer learning. Nat. Commun. 10, 2903 (2019).
Buterez, D., Janet, J. P., Kiddle, S. J., Oglic, D. & Lió, P. Transfer learning with graph neural networks for improved molecular property prediction in the multi-fidelity setting. Nat. Commun. 15, 1517 (2024).
Allen, A. E. A. et al. Learning together: towards foundation models for machine learning interatomic potentials with meta-learning. NPJ Comput. Mater. 10, 154 (2024).
Messerly, M. et al. Multi-fidelity learning for interatomic potentials: low-level forces and high-level energies are all you need. Mach. Learn.: Sci. Technol. 6, 035066 (2025).
Zubatyuk, R., Smith, J. S., Leszczynski, J. & Isayev, O. Accurate and transferable multitask prediction of chemical properties with an atoms-in-molecules neural network. Sci. Adv. 5, eaav6490 (2019).
Anstine, D. M., Zubatyuk, R. & Isayev, O. AIMNet2: a neural network potential to meet your neutral, charged, organic and elemental-organic needs. Chem. Sci. 16, 10228–10244 (2025).
Yao, K., Herr, J. E., Toth, D. W., Mckintyre, R. & Parkhill, J. The TensorMol-0.1 model chemistry: a neural network augmented with long-range physics. Chem. Sci. 9, 2261–2269 (2018).
Karwounopoulos, J. et al. Evaluation of machine learning/molecular mechanics end-state corrections with mechanical embedding to calculate relative protein-ligand binding free energies. J. Chem. Theory Comput. 21, 967–977 (2025).
Levine, D. S. et al. The Open Molecules 2025 (OMol25) dataset, evaluations and models. Preprint at https://arxiv.org/abs/2505.08762 (2025).
Thölke, P. & Fabritiis, G. D. TorchMD-NET: equivariant transformers for neural network based molecular potentials. Preprint at https://arxiv.org/abs/2202.02541 (2022).
Vaswani, A. et al. Attention is all you need. In Proc. Advances in Neural Information Processing Systems, Vol. 30, 5998–6008 (Curran Associates, Inc., 2017).
Tay, Y., Dehghani, M., Bahri, D. & Metzler, D. Efficient transformers: a survey. ACM Comput. Surv. 55, 1–28 (2023).
Frank, J. T., Unke, O. T. & Müller, K.-R. So3krates: equivariant attention for interactions on arbitrary length-scales in molecular systems. In Advances in Neural Information Processing Systems, Vol. 35, 29400–29413 (Curran Associates, Inc., 2022)
Qu, E. & Krishnapriyan, A. S. The importance of being scalable: improving the speed and accuracy of neural network interatomic potentials across chemical domains. In Advances in Neural Information Processing Systems, Vol. 37, 139030–139053 (Curran Associates, Inc., 2024).
Leimeroth, N., Erhard, L. C., Albe, K. & Rohrer, J. Machine-learning interatomic potentials from a users perspective: a comparison of accuracy, speed and data efficiency. Preprint at https://arxiv.org/abs/2505.02503 (2025).
Park, Y., Kim, J., Hwang, S. & Han, S. Scalable parallel algorithm for graph neural network interatomic potentials in molecular dynamics simulations. J. Chem. Theory Comput. 20, 4857–4868 (2024).
Zubatyuk, R. et al. AQuaRef: machine learning accelerated quantum refinement of protein structures. Nat. Commun. 16, 9224 (2025).
Accelerate Drug and Material Discovery with New Math Library NVIDIA cuEquivariance. NVIDIA Technical Blog (18 November 2024); https://developer.nvidia.com/blog/accelerate-drug-and-material-discovery-with-new-math-library-nvidia-cuequivariance/
Amin, I., Raja, S. & Krishnapriyan, A. Towards fast, specialized machine learning force fields: distilling foundation models via energy hessians. Preprint at https://arxiv.org/abs/2501.09009 (2025).
Matin, S. et al. Ensemble knowledge distillation for machine learning interatomic potentials. Preprint at https://arxiv.org/abs/2503.14293 (2025).
Senn, H. M. & Thiel, W. QM/MM methods for biomolecular systems. Angew. Chem. Int. Ed. 48, 1198–1229 (2009).
Lahey, S.-L. J. & Rowley, C. N. Simulating protein-ligand binding with neural network potentials. Chem. Sci. 11, 2362–2368 (2020).
Gastegger, M., Schütt, K. T. & Müller, K.-R. Machine learning of solvent effects on molecular spectra and reactions. Chem. Sci. 12, 11473–11483 (2021).
Sabanés Zariquiey, F. et al. Enhancing protein-ligand binding affinity predictions using neural network potentials. J. Chem. Inf. Model. 64, 1481–1485 (2024).
Nováček, M. & Řezáč, J. PM6-ML: the synergy of semiempirical quantum chemistry and machine learning transformed into a practical computational method. J. Chem. Theory Comput. 21, 678–690 (2025).
Valsson, Í et al. Narrowing the gap between machine learning scoring functions and free energy perturbation using augmented data. Commun. Chem. 8, 41 (2025).
Galvelis, R., Doerr, S., Damas, J. M., Harvey, M. J. & De Fabritiis, G. A scalable molecular force field parameterization method based on density functional theory and quantum-level machine learning. J. Chem. Inf. Model. 59, 3485–3493 (2019).
Tayfuroglu, O., Zengin, I. N., Koca, M. S. & Kocak, A. DeepConf: leveraging ANI-ML potentials for exploring local minima with application to bioactive conformations. J. Chem. Inf. Model. 65, 2818–2833 (2025).
Baillif, B., Cole, J., Giangreco, I., McCabe, P. & Bender, A. Applying atomistic neural networks to bias conformer ensembles towards bioactive-like conformations. J. Cheminformatics 15, 124 (2023).
Pan, X. et al. MolTaut: a tool for the rapid generation of favorable tautomer in aqueous solution. J. Chem. Inf. Model. 63, 1833–1840 (2023).
Han, F. et al. Distribution of bound conformations in conformational ensembles for X-ray ligands predicted by the ANI-2X machine learning potential. J. Chem. Inf. Model. 63, 6608–6618 (2023).
Berenger, F. & Tsuda, K. An ANI-2 enabled open-source protocol to estimate ligand strain after docking. J. Comput. Chem. 46, e27478 (2025).
Maestro (Schrödinger); https://www.schrodinger.com/platform/products/maestro/
Accelerate your chemistry & materials research (SCM); https://www.scm.com/
BIOVIA (Dassault Systèmes); https://www.3ds.com/products/biovia
Dral, P. O. et al. MLatom 3: a platform for machine learning-enhanced computational chemistry simulations and workflows. J. Chem. Theory Comput. 20, 1193–1213 (2024).
Zhao, Q. et al. Comprehensive exploration of graphically defined reaction spaces. Sci. Data 10, 145 (2023).
Liu, Z., Moroz, Y. S. & Isayev, O. The challenge of balancing model sensitivity and robustness in predicting yields: a benchmarking study of amide coupling reactions. Chem. Sci. 14, 10835–10846 (2023).
Revolutionizing AI-Driven Material Discovery Using NVIDIA ALCHEMI. NVIDIA Technical Blog (18 November 2025); https://developer.nvidia.com/blog/revolutionizing-ai-driven-material-discovery-using-nvidia-alchemi
Spotlight: Shell Accelerates CO2 Storage Modeling 100,000x Using NVIDIA PhysicsNeMo. NVIDIA Technical Blog (9 September 2024); https://developer.nvidia.com/blog/spotlight-shell-accelerates-co2-storage-modeling-100000x-using-nvidia-physicsnemo
St. John, P. S. et al. BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery. Preprint at https://arxiv.org/abs/2411.10548 (2024).
Boiko, D. A., Reschützegger, T., Sanchez-Lengeling, B., Blau, S. M. & Gomes, G. Advancing molecular machine learning representations with stereoelectronics-infused molecular graphs. Nat. Mach. Intell. 7, 771–781 (2025).
Qiao, Z., Welborn, M., Anandkumar, A., Manby, F. R. & Miller, T. F. III OrbNet: deep learning for quantum chemistry using symmetry-adapted atomic-orbital features. J. Chem. Phys. 153, 124111 (2020).
Qiao, Z. et al. Informing geometric deep learning with electronic interactions to accelerate quantum chemistry. Proc. Natl Acad. Sci. USA 119, e2205221119 (2022).
Kang, B. S. et al. OrbitAll: a unified quantum mechanical representation deep learning framework for all molecular systems. Preprint at https://arxiv.org/abs/2507.03853 (2025).
Kabylda, A. et al. Molecular simulations with a pretrained neural network and universal pairwise force fields. J. Am. Chem. Soc. 147, 33723–33734 (2025).
Releases · ACEsuit/mace. GitHub https://github.com/ACEsuit/mace/releases (accessed 17 September 2025).
Unke, O. T. et al. SpookyNet: learning force fields with electronic degrees of freedom and nonlocal effects. Nat. Commun. 12, 7273 (2021).
Kalita, B. et al. AIMNet2-NSE: a transferable reactive neural network potential for open-shell chemistry. Preprint at ChemRxiv https://doi.org/10.26434/chemrxiv-2025-kdg6n (2025).
Zubatyuk, R., Smith, J. S., Nebgen, B. T., Tretiak, S. & Isayev, O. Teaching a neural network to attach and detach electrons from molecules. Nat. Commun. 12, 4870 (2021).
Gelžinytė, E., Öeren, M., Segall, M. D. & Csányi, G. Transferable machine learning interatomic potential for bond dissociation energy prediction of drug-like molecules. J. Chem. Theory Comput. 20, 164–177 (2024).
Yang, Y., Zhang, S., Ranasinghe, K. D., Isayev, O. & Roitberg, A. E. Machine learning of reactive potentials. Annu. Rev. Phys. Chem. 75, 371–395 (2024).
Wang, L.-P. et al. Discovering chemistry with an ab initio nanoreactor. Nat. Chem. 6, 1044–1048 (2014).
Chen, B. W. J., Zhang, X. & Zhang, J. Accelerating explicit solvent models of heterogeneous catalysts with machine learning interatomic potentials. Chem. Sci. 14, 8338–8354 (2023).
Unke, O. T. & Meuwly, M. PhysNet: a neural network for predicting energies, forces, dipole moments and partial charges. J. Chem. Theory Comput. 15, 3678–3693 (2019).
Yu, H., Xu, Z., Qian, X., Qian, X. & Ji, S. Efficient and equivariant graph networks for predicting quantum Hamiltonian. In Proceedings of the 40th International Conference on Machine Learning, Vol. 202, 40412–40424 (PMLR, 2023).
Luise, G. et al. Accurate and scalable exchange-correlation with deep learning. Preprint at https://arxiv.org/abs/2506.14665 (2025).
Froitzheim, T., Müller, M., Hansen, A. & Grimme, S. G-xTB: a general-purpose extended tight-binding electronic structure method for the elements H to Lr (Z = 1–103). Preprint at ChemRxiv https://doi.org/10.26434/chemrxiv-2025-bjxvt (2025).
Bannwarth, C., Ehlert, S. & Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 15, 1652–1671 (2019).
Choi, J., Nam, G., Choi, J. & Jung, Y. A perspective on foundation models in chemistry. JACS Au 5, 1499–1518 (2025).
Eastman, P., Pritchard, B. P., Chodera, J. D. & Markland, T. E. Nutmeg and SPICE: models and data for biomolecular machine learning. J. Chem. Theory Comput. 20, 8583–8593 (2024).
Schreiner, M., Bhowmik, A., Vegge, T., Busk, J. & Winther, O. Transition1x—a dataset for building generalizable reactive machine learning potentials. Sci. Data 9, 779 (2022).
Plé, T. et al. A foundation model for accurate atomistic simulations in drug design. Preprint at ChemRxiv https://doi.org/10.26434/chemrxiv-2025-f1hgn-v3 (2025).
Chiang, Y. et al. MLIP Arena: advancing fairness and transparency in machine learning interatomic potentials through an open and accessible benchmark platform. Preprint at https://arxiv.org/abs/2509.20630 (2025).
FAIR Chemistry Leaderboard—a Hugging Face Space by Facebook https://huggingface.co/spaces/facebook/fairchem_leaderboard (accessed 17 September 2025).
Schaaf, L., Fako, E., De, S., Schäfer, A. & Csányi, G. Accurate energy barriers for catalytic reaction pathways: an automatic training protocol for machine learning force fields. npj Comput Mater 9, 180 (2023).
Kouw, W. M. & Loog, M. An introduction to domain adaptation and transfer learning. Preprint at https://arxiv.org/abs/1812.11806 (2019).
Pfeiffer, J., Ruder, S., Vulić, I. & Ponti, E. M. Modular deep learning. Preprint at https://arxiv.org/abs/2302.11529 (2024).
Chen, X., Wang, S., Fu, B., Long, M. & Wang, J. Catastrophic forgetting meets negative transfer: batch spectral shrinkage for safe transfer learning. In Proc. Advances in Neural Information Processing Systems, Vol. 32, 1908–1918 (Curran Associates, Inc., 2019).
Kirkpatrick, J. et al. Overcoming catastrophic forgetting in neural networks. Proc. Natl Acad. Sci. USA 114, 3521–3526 (2017).
Hüllermeier, E. & Waegeman, W. Aleatoric and epistemic uncertainty in machine learning: an introduction to concepts and methods. Mach. Learn. 110, 457–506 (2021).
Kulichenko, M. et al. Data generation for machine learning interatomic potentials and beyond. Chem. Rev. 124, 13681–13714 (2024).
Kulichenko, M. et al. Uncertainty-driven dynamics for active learning of interatomic potentials. Nat. Comput. Sci. 3, 230–239 (2023).
Glavatskikh, M., Leguy, J., Hunault, G., Cauchy, T. & Da Mota, B. Dataset’s chemical diversity limits the generalizability of machine learning predictions. J. Cheminformatics 11, 69 (2019).
Korth, M. & Grimme, S. Mindless’ DFT benchmarking. J. Chem. Theory Comput. 5, 993–1003 (2009).
Gould, T., Chan, B., Dale, S. G. & Vuckovic, S. Identifying and embedding transferability in data-driven representations of chemical space. Chem. Sci. 15, 11122–11133 (2024).
Bolhuis, P. G., Chandler, D., Dellago, C. & Geissler, P. L. TRANSITION PATH SAMPLING: throwing ropes over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 53, 291–318 (2002).
Jung, H., Okazaki, K. & Hummer, G. Transition path sampling of rare events by shooting from the top. J. Chem. Phys. 147, 152716 (2017).
Anstine, D. M. et al. AIMNet2-Rxn: a machine learned potential for generalized reaction modeling on a millions-of-pathways scale. Preprint at ChemRxiv https://doi.org/10.26434/chemrxiv-2025-hpdmg (2025).
Poongavanam, V. et al. Conformational sampling of macrocyclic drugs in different environments: can we find the relevant conformations?. ACS Omega 3, 11742–11757 (2018).
Witek, J. et al. Kinetic models of cyclosporin A in polar and apolar environments reveal multiple congruent conformational states. J. Chem. Inf. Model. 56, 1547–1562 (2016).
Kamenik, A. S., Lessel, U., Fuchs, J. E., Fox, T. & Liedl, K. R. Peptidic macrocycles - conformational sampling and thermodynamic characterization. J. Chem. Inf. Model. 58, 982–992 (2018).
Shrestha, U. R., Smith, J. C. & Petridis, L. Full structural ensembles of intrinsically disordered proteins from unbiased molecular dynamics simulations. Commun. Biol. 4, 243 (2021).
Potoyan, D. A. & Papoian, G. A. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. J. Am. Chem. Soc. 133, 7405–7415 (2011).
Appadurai, R., Nagesh, J. & Srivastava, A. High resolution ensemble description of metamorphic and intrinsically disordered proteins using an efficient hybrid parallel tempering scheme. Nat. Commun. 12, 958 (2021).
Morrow, J. D., Gardner, J. L. A. & Deringer, V. L. How to validate machine-learned interatomic potentials. J. Chem. Phys. 158, 121501 (2023).
Vassilev-Galindo, V., Fonseca, G., Poltavsky, I. & Tkatchenko, A. Challenges for machine learning force fields in reproducing potential energy surfaces of flexible molecules. J. Chem. Phys. 154, 094119 (2021).
Xin, H., Kitchin, J. R. & Kulik, H. J. Towards agentic science for advancing scientific discovery. Nat. Mach. Intell. 7, 1373–1375 (2025).
Aspuru-Guzik, A. & Bernales, V. The rise of agents: computational chemistry is ready for (R)evolution. Polyhedron 281, 117707 (2025).
Acknowledgements
We acknowledge support by the National Science Foundation (NSF) through the Center for Computer-Assisted Synthesis (C-CAS) CHE-2202693 award and the Office of Naval Research (ONR) through the Energetic Materials Program (MURI grant no. N00014-21-1-2476). This research is part of the Frontera computing project at the Texas Advanced Computing Center. Frontera is made possible by NSF award OAC-1818253.
Author information
Authors and Affiliations
Contributions
All authors participated in the writing of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Abdulrahman Aldossary and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kalita, B., Gokcan, H. & Isayev, O. Machine learning interatomic potentials at the centennial crossroads of quantum mechanics. Nat Comput Sci 5, 1120–1132 (2025). https://doi.org/10.1038/s43588-025-00930-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-025-00930-6