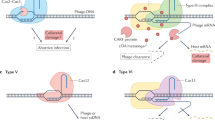
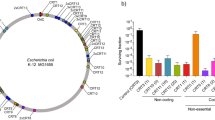
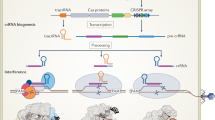
Abstract
Technologies derived from the CRISPR (clustered regularly interspaced short palindromic repeats)–Cas immune system of prokaryotes have revolutionized our ability to cleave and modify target nucleic acid sequences. In addition to the use of CRISPR–Cas tools for the editing of human genes, they can also be designed to target pathogenic and commensal bacteria that colonize the body, offering new pathways for the treatment of infections and microbiome modulation. In this Review, we explore how the CRISPR–Cas toolbox can be engineered to kill or modify specific bacteria. We discuss DNA-targeting and RNA-targeting strategies, outlining how these can be applied to disarm bacteria by removing, modifying or silencing specific genes. Furthermore, we examine the delivery of CRISPR–Cas tools by bacteriophages and through conjugation and explore intracellular barriers to CRISPR–Cas tool maintenance and expression. Finally, we highlight therapeutic opportunities in the treatment of infectious diseases and for the modification of the microbiome, outlining progress and challenges in translating these approaches into clinical applications.
Key points
-
CRISPR (clustered regularly interspaced short palindromic repeats)–Cas systems can be designed as tools to kill or modify target bacteria, for example, by targeting antibiotic resistance genes, virulence factors or genes involved in microbiome-related diseases.
-
CRISPR–Cas tools can be delivered to target bacteria by phage particles or plasmid conjugation.
-
Delivery of CRISPR–Cas therapeutics requires engineering to enhance efficiency, adapt the host range to the target strains and bypass bacterial defence mechanisms.
-
First clinical trials have demonstrated the safety of Cas-armed phages.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Okeke, I. N. et al. The scope of the antimicrobial resistance challenge. Lancet 403, 2426–2438 (2024).
Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71 (2021).
Hou, K. et al. Microbiota in health and diseases. Signal. Transduct. Target. Ther. 7, 135 (2022).
Pirnay, J.-P. et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: a multicentre, multinational, retrospective observational study. Nat. Microbiol. 9, 1434–1453 (2024).
Lei, M., Jayaraman, A., Deventer, J. A. V. & Lee, K. Engineering selectively targeting antimicrobial peptides. Annu. Rev. Biomed. Eng. 23, 339–357 (2021).
Altae-Tran, H. et al. Uncovering the functional diversity of rare CRISPR-Cas systems with deep terascale clustering. Science 382, eadi1910 (2023).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
Nussenzweig, P. M. & Marraffini, L. A. Molecular mechanisms of CRISPR-Cas immunity in bacteria. Annu. Rev. Genet. 54, 93–120 (2020).
Rodrigues, M., McBride, S. W., Hullahalli, K., Palmer, K. L. & Duerkop, B. A. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant Enterococci. Antimicrob. Agents Chemother. 63, e01454-19 (2019).
Bikard, D. et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150 (2014).
Citorik, R. J., Mimee, M. & Lu, T. K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145 (2014).
Lam, K. N. et al. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep. 37, 109930 (2021).
Park, J. Y. et al. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 7, 44929 (2017).
Ram, G., Ross, H. F., Novick, R. P., Rodriguez-Pagan, I. & Jiang, D. Conversion of staphylococcal pathogenicity islands to CRISPR-carrying antibacterial agents that cure infections in mice. Nat. Biotechnol. 36, 971–976 (2018).
Neil, K. et al. High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing. Mol. Syst. Biol. 17, e10335 (2021).
Jin, W.-B. et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell 185, 547–562.e22 (2022).
Zhou, Y. et al. Exploiting a conjugative endogenous CRISPR-Cas3 system to tackle multidrug-resistant Klebsiella pneumoniae. eBioMedicine 88, 104445 (2023).
Selle, K. et al. In vivo targeting of clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio 11, e00019-20 (2020).
Gencay, Y. E. et al. Engineered phage with antibacterial CRISPR–Cas selectively reduce E. coli burden in mice. Nat. Biotechnol. 42, 265–274 (2023).
Cui, L. & Bikard, D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 44, 4243–4251 (2016).
Csörgő, B. et al. A compact Cascade–Cas3 system for targeted genome engineering. Nat. Methods 17, 1183–1190 (2020).
Vercoe, R. B. et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9, e1003454 (2013).
Bernheim, A., Bikard, D., Touchon, M. & Rocha, E. P. C. A matter of background: DNA repair pathways as a possible cause for the sparse distribution of CRISPR-Cas systems in bacteria. Philos. Trans. R. Soc. B Biol. Sci. 374, 20180088 (2019).
Vialetto, E. et al. Systematic interrogation of CRISPR antimicrobials in Klebsiella pneumoniae reveals nuclease-, guide- and strain-dependent features influencing antimicrobial activity. Nucleic Acids Res. 52, 6079–6091 (2024).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Stella, G. & Marraffini, L. Type III CRISPR-Cas: beyond the Cas10 effector complex. Trends Biochem. Sci. 49, 28–37 (2024).
Rostøl, J. T. et al. The Card1 nuclease provides defence during type III CRISPR immunity. Nature 590, 624–629 (2021).
Marraffini, L. A. & Sontheimer, E. J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 (2010).
Pyenson, N. C., Gayvert, K., Varble, A., Elemento, O. & Marraffini, L. A. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22, 343–353.e3 (2017).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, eaaf5573 (2016).
Vialetto, E. et al. A target expression threshold dictates invader defense and prevents autoimmunity by CRISPR-Cas13. Cell Host Microbe 30, 1151–1162.e6 (2022).
Kiga, K. et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 11, 2934 (2020).
Dmytrenko, O. et al. Cas12a2 elicits abortive infection through RNA-triggered destruction of dsDNA. Nature 613, 588–594 (2023).
Selle, K., Klaenhammer, T. R. & Barrangou, R. CRISPR-based screening of genomic island excision events in bacteria. Proc. Natl Acad. Sci. USA 112, 8076–8081 (2015).
Wang, P. et al. Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J. Antimicrob. Chemother. 74, 2559–2565 (2019).
Yosef, I., Manor, M., Kiro, R. & Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 112, 7267–7272 (2015).
Long, T.-F. et al. Innovative delivery system combining CRISPR-Cas12f for combatting antimicrobial resistance in gram-negative bacteria. ACS Synth. Biol. 13, 1831–1841 (2024).
Paterson, D. L. & Bonomo, R. A. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686 (2005).
Tagliaferri, T. L. et al. Exploring the potential of CRISPR-Cas9 under challenging conditions: facing high-copy plasmids and counteracting beta-lactam resistance in clinical strains of Enterobacteriaceae. Front. Microbiol. 11, 578 (2020).
Wongpayak, P., Meesungnoen, O., Saejang, S. & Subsoontorn, P. A highly effective and self-transmissible CRISPR antimicrobial for elimination of target plasmids without antibiotic selection. PeerJ 9, e11996 (2021).
Dong, H., Xiang, H., Mu, D., Wang, D. & Wang, T. Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int. J. Antimicrob. Agents 53, 1–8 (2019).
Li, P. et al. Targeted elimination of bla NDM-5 gene in Escherichia coli by conjugative CRISPR-Cas9 system. Infect. Drug Resist. 15, 1707–1716 (2022).
Walker-Sünderhauf, D. et al. Removal of AMR plasmids using a mobile, broad host-range CRISPR-Cas9 delivery tool. Microbiology 169, 001334 (2023).
Bakkeren, E. et al. Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 573, 276–280 (2019).
Quinones-Olvera, N. et al. Diverse and abundant phages exploit conjugative plasmids. Nat. Commun. 15, 3197 (2024).
Jurėnas, D., Fraikin, N., Goormaghtigh, F. & Van Melderen, L. Biology and evolution of bacterial toxin–antitoxin systems. Nat. Rev. Microbiol. 20, 335–350 (2022).
Reuter, A. et al. Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity. Nucleic Acids Res. 49, 3584–3598 (2021).
Benz, F. et al. Type IV-A3 CRISPR-Cas systems drive inter-plasmid conflicts by acquiring spacers in trans. Cell Host Microbe 32, 875–886.e9 (2024).
Mamontov, V. et al. Persistence of plasmids targeted by CRISPR interference in bacterial populations. Proc. Natl Acad. Sci. USA 119, e2114905119 (2022).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Jiang, Y. et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015).
Reisch, C. R. & Prather, K. L. J. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci. Rep. 5, 15096 (2015).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Brödel, A. K. et al. In situ targeted base editing of bacteria in the mouse gut. Nature 632, 877–884 (2024).
Guzmán-Herrador, D. L., Fernández-Gómez, A., Depardieu, F., Bikard, D. & Llosa, M. In vivo delivery of functional Cas:DNA nucleoprotein complexes into recipient bacteria through a type IV secretion system. Proc. Natl Acad. Sci. USA 121, e2408509121 (2024).
Nethery, M. A., Hidalgo-Cantabrana, C., Roberts, A. & Barrangou, R. CRISPR-based engineering of phages for in situ bacterial base editing. Proc. Natl Acad. Sci. USA 119, e2206744119 (2022).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Gelsinger, D. R. et al. Bacterial genome engineering using CRISPR-associated transposases. Nat. Protoc. 19, 752–790 (2024).
Rubin, B. E. et al. Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 7, 34–47 (2022).
Bikard, D. et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437 (2013).
Qi, L. S. et al. Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Luo, M. L., Mullis, A. S., Leenay, R. T. & Beisel, C. L. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681 (2015).
Kim, S. K. et al. Efficient transcriptional gene repression by type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth. Biol. 6, 1273–1282 (2017).
Vigouroux, A. & Bikard, D. CRISPR tools to control gene expression in bacteria. Microbiol. Mol. Biol. Rev. 84, e00077-19 (2020).
Zebec, Z., Manica, A., Zhang, J., White, M. F. & Schleper, C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 42, 5280–5288 (2014).
Adler, B. A. et al. Genome-wide characterization of diverse bacteriophages enabled by RNA-binding CRISPRi. Nat. Microbiol. 10, 694–709 (2025).
Charles, E. J. et al. Engineering improved Cas13 effectors for targeted post-transcriptional regulation of gene expression. Preprint at bioRxiv https://doi.org/10.1101/2021.05.26.445687 (2021).
Kim, G. et al. Tunable translation-level CRISPR interference by dCas13 and engineered gRNA in bacteria. Nat. Commun. 15, 5319 (2024).
Madigan, V., Zhang, F. & Dahlman, J. E. Drug delivery systems for CRISPR-based genome editors. Nat. Rev. Drug Discov. 22, 875–894 (2023).
Bikard, D., Hatoum-Aslan, A., Mucida, D. & Marraffini, L. A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12, 177–186 (2012).
Gomaa, A. A. et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5, e00928-13 (2014).
Shkoporov, A. N. et al. Viral biogeography of the mammalian gut and parenchymal organs. Nat. Microbiol. 7, 1301–1311 (2022).
López-Pérez, M., Haro-Moreno, J. M., Gonzalez-Serrano, R., Parras-Moltó, M. & Rodriguez-Valera, F. Genome diversity of marine phages recovered from Mediterranean metagenomes: size matters. PLoS Genet. 13, e1007018 (2017).
Kauffman, K. M. et al. Resolving the structure of phage–bacteria interactions in the context of natural diversity. Nat. Commun. 13, 372 (2022).
Bertozzi Silva, J., Storms, Z. & Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363, fnw002 (2016).
Buffet, A., Rocha, E. P. C. & Rendueles, O. Nutrient conditions are primary drivers of bacterial capsule maintenance in Klebsiella. Proc. R. Soc. B Biol. Sci. 288, 20202876 (2021).
Klumpp, J., Dunne, M. & Loessner, M. J. A perfect fit: bacteriophage receptor-binding proteins for diagnostic and therapeutic applications. Curr. Opin. Microbiol. 71, 102240 (2023).
Latka, A. et al. Engineering the modular receptor-binding proteins of Klebsiella phages switches their capsule serotype specificity. mBio 12, e00455-21 (2021).
Ando, H., Lemire, S., Pires, D. P. & Lu, T. K. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 1, 187–196 (2015).
Apjok, G. et al. Characterization of antibiotic resistomes by reprogrammed bacteriophage-enabled functional metagenomics in clinical strains. Nat. Microbiol. 8, 410–423 (2023).
Cunliffe, T. G., Parker, A. L. & Jaramillo, A. Pseudotyping bacteriophage P2 tail fibers to extend the host range for biomedical applications. ACS Synth. Biol. 10, 3207–3215 (2022).
Dunne, M. et al. Reprogramming bacteriophage host range through structure-guided design of chimeric receptor binding proteins. Cell Rep. 29, 1336–1350.e4 (2019).
Lam, C. N. et al. A tail fiber engineering platform for improved bacterial transduction-based diagnostic reagents. ACS Synth. Biol. 10, 1292–1299 (2021).
Mahichi, F., Synnott, A. J., Yamamichi, K., Osada, T. & Tanji, Y. Site‐specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol. Lett. 295, 211–217 (2009).
Yosef, I., Goren, M. G., Globus, R., Molshanski-Mor, S. & Qimron, U. Extending the host range of bacteriophage particles for DNA transduction. Mol. Cell 66, 721–728.e3 (2017).
Montag, D., Schwarz, H. & Henning, U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J. Bacteriol. 171, 4378–4384 (1989).
Nobrega, F. L. et al. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 16, 760–773 (2018).
Ibarra-Chávez, R. Rebooting synthetic phage-inducible chromosomal islands: one method to forge them all. Biodes. Res. 2020, 5783064 (2020).
Huss, P., Meger, A., Leander, M., Nishikawa, K. K. & Raman, S. Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning. eLife 10, e63775 (2021).
Liang, J., Zhang, H., Tan, Y. L., Zhao, H. & Ang, E. L. Directed evolution of replication-competent double-stranded DNA bacteriophage toward new host specificity. ACS Synth. Biol. 11, 634–643 (2022).
Yehl, K. et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179, 459–469.e9 (2019).
Liyanagedera, S. B. W. et al. SpyPhage: a cell-free TXTL platform for rapid engineering of targeted phage therapies. ACS Synth. Biol. 11, 3330–3342 (2022).
Levrier, A. et al. PHEIGES: all-cell-free phage synthesis and selection from engineered genomes. Nat. Commun. 15, 2223 (2024).
Favor, A. H., Llanos, C. D., Youngblut, M. D. & Bardales, J. A. Optimizing bacteriophage engineering through an accelerated evolution platform. Sci. Rep. 10, 13981 (2020).
Nobrega, F. L. et al. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci. Rep. 6, 39235 (2016).
Kittleson, J. T., DeLoache, W., Cheng, H.-Y. & Anderson, J. C. Scalable plasmid transfer using engineered P1-based phagemids. ACS Synth. Biol. 1, 583–589 (2012).
Lu, T. K. & Collins, J. J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl Acad. Sci. USA 106, 4629–4634 (2009).
Tridgett, M., Ababi, M., Osgerby, A., Garcia, R. R. & Jaramillo, A. Engineering bacteria to produce pure phage-like particles for gene delivery. ACS Synth. Biol. 10, 107–114 (2021).
Westwater, C., Schofield, D. A., Schmidt, M. G., Norris, J. S. & Dolan, J. W. Development of a P1 phagemid system for the delivery of DNA into Gram-negative bacteria. Microbiology 148, 943–950 (2002).
Edgar, R., Friedman, N., Molshanski-Mor, S. & Qimron, U. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl. Environ. Microbiol. 78, 744–751 (2012).
Hsu, B. B. et al. In situ reprogramming of gut bacteria by oral delivery. Nat. Commun. 11, 5030 (2020).
Al-Shayeb, B. et al. Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431 (2020).
Cronan, J. E. Improved plasmid-based system for fully regulated off-to-on gene expression in Escherichia coli: application to production of toxic proteins. Plasmid 69, 81–89 (2013).
Ibarra-Chávez, R., Hansen, M. F., Pinilla-Redondo, R., Seed, K. D. & Trivedi, U. Phage satellites and their emerging applications in biotechnology. FEMS Microbiol. Rev. 45, fuab031 (2021).
de Sousa, J. A. M., Fillol-Salom, A., Penadés, J. R. & Rocha, E. P. C. Identification and characterization of thousands of bacteriophage satellites across bacteria. Nucleic Acids Res. 51, 2759–2777 (2023).
Chen, J. & Novick, R. P. Phage-mediated intergeneric transfer of toxin genes. Science 323, 139–141 (2009).
Caro, L. G. & Schnös, M. The attachment of the male-specific bacteriophage F1 to sensitive strains of Escherichia coli. Proc. Natl Acad. Sci. USA 56, 126–132 (1966).
Loeb, T. Isolation of a bacteriophage specific for the F+ and Hfr mating types of Escherichia coli K-12. Science 131, 932–933 (1960).
Paepe, M. D. et al. Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet. 7, e1002107 (2011).
Paepe, M. D. et al. Carriage of λ latent virus is costly for its bacterial host due to frequent reactivation in monoxenic mouse intestine. PLoS Genet. 12, e1005861 (2016).
Bernabéu-Gimeno, M. et al. Neutralizing antibodies after nebulized phage therapy in cystic fibrosis patients. Med 5, 1096–1111.e6 (2024).
Dedrick, R. M. et al. Phage therapy of mycobacterium infections: compassionate use of phages in 20 patients with drug-resistant mycobacterial disease. Clin. Infect. Dis. 76, 103–112 (2023).
Lourenço, M. et al. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401.e5 (2020).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl Acad. Sci. USA 110, 10771–10776 (2013).
Lu, T. K. & Collins, J. J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl Acad. Sci. USA 104, 11197–11202 (2007).
Chin, W. H. et al. Bacteriophages evolve enhanced persistence to a mucosal surface. Proc. Natl Acad. Sci. USA 119, e2116197119 (2022).
Ronda, C., Chen, S. P., Cabral, V., Yaung, S. J. & Wang, H. H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 16, 167–170 (2019).
Klümper, U. et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945 (2015).
Mazodier, P. & Davies, J. Gene transfer between distantly related bacteria. Annu. Rev. Genet. 25, 147–171 (1991).
Johnson, C. M. & Grossman, A. D. Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601 (2015).
Peters, J. M. et al. Enabling genetic analysis of diverse bacteria with mobile-CRISPRi. Nat. Microbiol. 4, 244–250 (2019).
Djermoun, S., Reuter, A., Derollez, E., Lesterlin, C. & Bigot, S. Reprogramming targeted-antibacterial-plasmids (TAPs) to achieve broad-host range antibacterial activity. Plasmid 126, 102680 (2023).
Hamilton, T. A. et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat. Commun. 10, 4544 (2019).
Ji, W. et al. Specific gene repression by CRISPRi system transferred through bacterial conjugation. ACS Synth. Biol. 3, 929–931 (2014).
Li, X. et al. Degradation of antibiotic resistance genes by VADER with CRISPR-Cas immunity. Appl. Environ. Microbiol. 89, e00053-23 (2023).
Ruotsalainen, P., Penttinen, R., Mattila, S. & Jalasvuori, M. Midbiotics: conjugative plasmids for genetic engineering of natural gut flora. Gut Microbes 10, 643–653 (2019).
Sheng, H. et al. Engineering conjugative CRISPR-Cas9 systems for the targeted control of enteric pathogens and antibiotic resistance. PLoS One 18, e0291520 (2023).
Song, Z. et al. Pathogen-specific bactericidal method mediated by conjugative delivery of CRISPR-Cas13a targeting bacterial endogenous transcripts. Microbiol. Spectr. 10, e0130022 (2022).
Simon, R., Priefer, U. & Pühler, A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1, 784–791 (1983).
Bradley, D. E., Taylor, D. E. & Cohen, D. R. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 143, 1466–1470 (1980).
Arutyunov, D. & Frost, L. S. F conjugation: back to the beginning. Plasmid 70, 18–32 (2013).
Bradley, D. E. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J. Gen. Microbiol. 130, 1489–1502 (1984).
Ishiwa, A. & Komano, T. The lipopolysaccharide of recipient cells is a specific receptor for PilV proteins, selected by shufflon DNA rearrangement, in liquid matings with donors bearing the R64 plasmid. Mol. Gen. Genet. 263, 159–164 (2000).
Neil, K., Allard, N., Grenier, F., Burrus, V. & Rodrigue, S. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl2 conjugative plasmid TP114. Commun. Biol. 3, 523 (2020).
Hamilton, T. A. et al. De novo synthesis of a conjugative system from human gut metagenomic data for targeted delivery of Cas9 antimicrobials. ACS Synth. Biol. 12, 3578–3590 (2023).
Fox, R. E., Zhong, X., Krone, S. M. & Top, E. M. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2, 1024–1039 (2008).
Benz, F. et al. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 15, 862–878 (2020).
Licht, T. R., Christensen, B. B., Krogfelt, K. A. & Molin, S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145, 2615–2622 (1999).
Christensen, B. B. et al. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64, 2247–2255 (1998).
Haagensen, J. A. J., Hansen, S. K., Johansen, T. & Molin, S. In situ detection of horizontal transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42, 261–268 (2002).
Neil, K., Allard, N. & Rodrigue, S. Molecular mechanisms influencing bacterial conjugation in the intestinal microbiota. Front. Microbiol. 12, 673260 (2021).
Allard, N., Collette, A., Paquette, J., Rodrigue, S. & Côté, J.-P. Systematic investigation of recipient cell genetic requirements reveals important surface receptors for conjugative transfer of IncI2 plasmids. Commun. Biol. 6, 1172 (2023).
Ishiwa, A. & Komano, T. Thin pilus PilV adhesins of plasmid R64 recognize specific structures of the lipopolysaccharide molecules of recipient cells. J. Bacteriol. 185, 5192–5199 (2003).
Low, W. W. et al. Mating pair stabilization mediates bacterial conjugation species specificity. Nat. Microbiol. 7, 1016–1027 (2022).
Robledo, M. et al. Targeted bacterial conjugation mediated by synthetic cell-to-cell adhesions. Nucleic Acids Res. 50, 12938–12950 (2022).
Allard, N., Neil, K., Grenier, F. & Rodrigue, S. The type IV pilus of plasmid TP114 displays adhesins conferring conjugation specificity and is important for DNA transfer in the mouse gut microbiota. Microbiol. Spectr. 10, e0230321 (2022).
Brouwer, M. S. M. et al. The shufflon of IncI1 plasmids is rearranged constantly during different growth conditions. Plasmid 102, 51–55 (2019).
Komano, T., Kubo, A. & Nisioka, T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 15, 1165–1172 (1987).
Dąbrowska, K. Phage therapy: what factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med. Res. Rev. 39, 2000–2025 (2019).
Caballero-Flores, G., Pickard, J. M. & Núñez, G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat. Rev. Microbiol. 21, 347–360 (2023).
Kristensen, N. B. et al. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 8, 52 (2016).
Georjon, H. & Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 21, 686–700 (2023).
Furuta, Y. & Kobayashi, I. Restriction-modification systems as mobile epigenetic elements. Madame Curie Bioscience Database (Landes Bioscience, 2013).
Loenen, W. A. M. & Raleigh, E. A. The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 42, 56–69 (2014).
Aparicio-Maldonado, C. et al. Class I DISARM provides anti-phage and anti-conjugation activity by unmethylated DNA recognition. Preprint at bioRxiv https://doi.org/10.1101/2021.12.28.474362 (2021).
Goldfarb, T. et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183 (2015).
Ofir, G. et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 3, 90–98 (2018).
Xiong, L. et al. A new type of DNA phosphorothioation-based antiviral system in archaea. Nat. Commun. 10, 1688 (2019).
Johnston, C. D. et al. Systematic evasion of the restriction-modification barrier in bacteria. Proc. Natl Acad. Sci. USA 116, 11454–11459 (2019).
Roberts, R. J., Vincze, T., Posfai, J. & Macelis, D. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015).
Dimitriu, T., Szczelkun, M. D. & Westra, E. R. Various plasmid strategies limit the effect of bacterial restriction-modification systems against conjugation. Nucleic Acids Res. 52, 12976–12986 (2024).
Roer, L., Aarestrup, F. M. & Hasman, H. The EcoKI type I restriction-modification system in Escherichia coli affects but is not an absolute barrier for conjugation. J. Bacteriol. 197, 337–342 (2015).
Maffei, E. et al. Systematic exploration of Escherichia coli phage-host interactions with the BASEL phage collection. PLoS Biol. 19, e3001424 (2021).
Zeng, X., Wu, Z., Zhang, Q. & Lin, J. A cotransformation method to identify a restriction-modification enzyme that reduces conjugation efficiency in Campylobacter jejuni. Appl. Environ. Microbiol. 84, e02004-18 (2018).
Suzuki, H. In Methylation — from DNA, RNA and Histones to Diseases and Treatment (ed. Dricu, A.) https://doi.org/10.5772/51691 (IntechOpen, 2012).
Vento, J. M. et al. A cell-free transcription-translation pipeline for recreating methylation patterns boosts DNA transformation in bacteria. Mol. Cell https://doi.org/10.1016/j.molcel.2024.06.003 (2024).
Emslander, Q. et al. Cell-free production of personalized therapeutic phages targeting multidrug-resistant bacteria. Cell Chem. Biol. 29, 1434–1445.e7 (2022).
Mayo-Muñoz, D., Pinilla-Redondo, R., Camara-Wilpert, S., Birkholz, N. & Fineran, P. C. Inhibitors of bacterial immune systems: discovery, mechanisms and applications. Nat. Rev. Genet. 25, 237–254 (2024).
Kudryavtseva, A. A. et al. Broadness and specificity: ArdB, ArdA, and Ocr against various restriction-modification systems. Front. Microbiol. 14, 1133144 (2023).
Dharmalingam, K., Revel, H. R. & Goldberg, E. B. Physical mapping and cloning of bacteriophage T4 anti-restriction endonuclease gene. J. Bacteriol. 149, 694–699 (1982).
Jaskólska, M., Adams, D. W. & Blokesch, M. Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae. Nature 604, 323–329 (2022).
Zongo, P. D. et al. An antiplasmid system drives antibiotic resistance gene integration in carbapenemase-producing Escherichia coli lineages. Nat. Commun. 15, 4093 (2024).
Liu, H. W. et al. DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage. Mol. Cell 82, 4727–4740.e6 (2022).
Tesson, F. et al. A comprehensive resource for exploring antiphage defense: DefensefFinder webservice,wiki and databases. Peer Community J. 4, e91 (2024).
Samuel, B., Mittelman, K., Croitoru, S. Y., Ben Haim, M. & Burstein, D. Diverse anti-defence systems are encoded in the leading region of plasmids. Nature 635, 186–192 (2024).
Bouet, J. Y., Nordström, K. & Lane, D. Plasmid partition and incompatibility — the focus shifts. Mol. Microbiol. 65, 1405–1414 (2007).
Ebersbach, G. & Gerdes, K. Plasmid segregation mechanisms. Annu. Rev. Genet. 39, 453–479 (2005).
Jiang, Y. et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 8, 15179 (2017).
Naduthodi, M. I. S., Barbosa, M. J. & van der Oost, J. Progress of CRISPR-Cas based genome editing in photosynthetic microbes. Biotechnol. J. 13, 1700591 (2018).
Rock, J. M. et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2, 16274 (2017).
Cui, L. et al. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 9, 1912 (2018).
Depardieu, F. & Bikard, D. Gene silencing with CRISPRi in bacteria and optimization of dCas9 expression levels. Methods 172, 61–75 (2020).
Rostain, W. et al. Cas9 off-target binding to the promoter of bacterial genes leads to silencing and toxicity. Nucleic Acids Res. 51, 3485–3496 (2023).
Kudla, G., Murray, A. W., Tollervey, D. & Plotkin, J. B. Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (2009).
Porse, A., Schou, T. S., Munck, C., Ellabaan, M. M. H. & Sommer, M. O. A. Biochemical mechanisms determine the functional compatibility of heterologous genes. Nat. Commun. 9, 522 (2018).
Tuller, T. et al. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 39, 4743–4755 (2011).
Uribe, R. V. et al. Bacterial resistance to CRISPR-Cas antimicrobials. Sci. Rep. 11, 17267 (2021).
Gomes, A. L. C. et al. Genome and sequence determinants governing the expression of horizontally acquired DNA in bacteria. ISME J. 14, 2347–2357 (2020).
Johns, N. I. et al. Metagenomic mining of regulatory elements enables programmable species-selective gene expression. Nat. Methods 15, 323–329 (2018).
Lamberte, L. E. et al. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat. Microbiol. 2, 16249 (2017).
Singh, K., Milstein, J. N. & Navarre, W. W. Xenogeneic silencing and its impact on bacterial genomes. Annu. Rev. Microbiol. 70, 199–213 (2016).
Schelling, M. A., Nguyen, G. T. & Sashital, D. G. CRISPR-Cas effector specificity and cleavage site determine phage escape outcomes. PLoS Biol. 21, e3002065 (2023).
Davidson, A. R. et al. Anti-CRISPRs: protein inhibitors of CRISPR-Cas systems. Annu. Rev. Biochem. 89, 309–332 (2020).
Castledine, M. et al. Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. eLife 11, e73679 (2022).
Wright, R. C. T., Friman, V.-P., Smith, M. C. M. & Brockhurst, M. A. Resistance evolution against phage combinations depends on the timing and order of exposure. mBio 10, e01652-19 (2019).
Averbuch, D. et al. Antimicrobial resistance in gram-negative rods causing bacteremia in hematopoietic stem cell transplant recipients: intercontinental prospective study of the infectious diseases working party of the European Bone Marrow Transplantation Group. Clin. Infect. Dis. 65, 1819–1828 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT05277350 (2023).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/study/NCT04191148 (2022).
Kim, P., Sanchez, A., Kime, J. & Ousterout, D. 1083. Phase 1b results of pharmacokinetics, pharmacodynamics, and safety for LBP-EC01, a CRISPR-Cas3 enhanced bacteriophage cocktail targeting escherichia coli that cause urinary tract infections. Open Forum Infect. Dis. 8, S633 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT05488340 (2024).
Kim, P. et al. Safety, pharmacokinetics, and pharmacodynamics of LBP-EC01, a CRISPR-Cas3-enhanced bacteriophage cocktail, in uncomplicated urinary tract infections due to Escherichia coli (ELIMINATE): the randomised, open-label, first part of a two-part phase 2 trial. Lancet Infect. Dis. 24, 1319–1332 (2024).
Sheitoyan-Pesant, C. et al. Clinical and healthcare burden of multiple recurrences of Clostridium difficile infection. Clin. Infect. Dis. 62, 574–580 (2016).
Johnson, S. et al. Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin. Infect. Dis. 73, e1029–e1044 (2021).
van Prehn, J. et al. European Society of Clinical Microbiology and Infectious Diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin. Microbiol. Infect. 27, S1–S21 (2021).
Umansky, A. A. & Fortier, L. C. The long and sinuous road to phage-based therapy of Clostridioides difficile infections. Front. Med. 10, 1259427 (2023).
Fitz-Gibbon, S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 (2013).
Mei, Z. et al. Strain-specific gut microbial signatures in type 2 diabetes identified in a cross-cohort analysis of 8,117 metagenomes. Nat. Med. 30, 2265–2276 (2024).
Leveau, A., Canadas Blasco, I., Mathieu, A. & Decrulle, A. Phage-derived particles for in situ delivery of DNA payload into C. acnes population. US Patent US20240254496A1 (2024).
Freedman, S. B., van de Kar, N. C. A. J. & Tarr, P. I. Shiga toxin-producing escherichia coli and the hemolytic-uremic syndrome. N. Engl. J. Med. 389, 1402–1414 (2023).
Galtier, M. et al. Treatment of STEC infection via CRISPR-Cas targeted cleavage of the Shiga toxin gene in animal models. Preprint at bioRxiv https://doi.org/10.1101/2025.02.28.640725 (2025).
Pacia, D. M., Brown, B. L., Minssen, T. & Darrow, J. J. CRISPR-phage antibacterials to address the antibiotic resistance crisis: scientific, economic, and regulatory considerations. J. Law Biosci. 11, lsad030 (2024).
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K. & Schooley, R. T. Phage therapy: from biological mechanisms to future directions. Cell 186, 17–31 (2023).
López-Igual, R., Bernal-Bayard, J., Rodríguez-Patón, A., Ghigo, J. M. & Mazel, D. Engineered toxin–intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat. Biotechnol. 37, 755–760 (2019).
Charbonneau, M. R., Isabella, V. M., Li, N. & Kurtz, C. B. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 11, 1738 (2020).
Ozdemir, T., Fedorec, A. J. H., Danino, T. & Barnes, C. P. Synthetic biology and engineered live biotherapeutics: toward increasing system complexity. Cell Syst. 7, 5–16 (2018).
Sheridan, C. The world’s first CRISPR therapy is approved: who will receive it? Nat. Biotechnol. 42, 3–4 (2023).
Tou, C. J. & Kleinstiver, B. P. Programmable RNA-guided enzymes for next-generation genome editing. Nature 630, 827–828 (2024).
Calvo-Villamañán, A. et al. On-target activity predictions enable improved CRISPR–dCas9 screens in bacteria. Nucleic Acids Res. 48, e64 (2020).
Gutierrez, B., Ng, J. W., Cui, L., Becavin, C. & Bikard, D. Genome-wide CRISPR-Cas9 screen in E. coli identifies design rules for efficient targeting. Preprint at bioRxiv https://doi.org/10.1101/308148 (2018).
Guo, J. et al. Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Res. 46, 7052–7069 (2018).
Hawkins, J. S. et al. Mismatch-CRISPRi reveals the co-varying expression-fitness relationships of essential genes in Escherichia coli and Bacillus subtilis. Cell Syst. 11, 523–535.e9 (2020).
Vigouroux, A., Oldewurtel, E., Cui, L., Bikard, D. & van Teeffelen, S. Tuning dCas9’s ability to block transcription enables robust, noiseless knockdown of bacterial genes. Mol. Syst. Biol. 14, e7899 (2018).
Rottinghaus, A. G., Vo, S. & Moon, T. S. Computational design of CRISPR guide RNAs to enable strain-specific control of microbial consortia. Proc. Natl Acad. Sci. USA 120, e2213154120 (2022).
Yu, Y. et al. Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration. Genome Biol. 25, 13 (2024).
Moreb, E. A. et al. Managing the SOS response for enhanced CRISPR-Cas-based recombineering in E. coli through transient inhibition of host RecA activity. ACS Synth. Biol. 6, 2209–2218 (2017).
Rottinghaus, A. G., Ferreiro, A., Fishbein, S. R. S., Dantas, G. & Moon, T. S. Genetically stable CRISPR-based kill switches for engineered microbes. Nat. Commun. 13, 672 (2022).
Forier, K. et al. Lipid and polymer nanoparticles for drug delivery to bacterial biofilms. J. Control. Rel. 190, 607–623 (2014).
Lee, H. W., Kharel, S. & Loo, S. C. J. Lipid-coated hybrid nanoparticles for enhanced bacterial biofilm penetration and antibiofilm efficacy. ACS Omega 7, 35814–35824 (2022).
Moreira, L. et al. Liposome delivery of nucleic acids in bacteria: toward in vivo labeling of human microbiota. ACS Infect. Dis. 8, 1218–1230 (2022).
Pereira, S. et al. Lipoplexes to deliver oligonucleotides in gram-positive and gram-negative bacteria: towards treatment of blood infections. Pharmaceutics 13, 989 (2021).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Wagner, V., Dullaart, A., Bock, A.-K. & Zweck, A. The emerging nanomedicine landscape. Nat. Biotechnol. 24, 1211–1217 (2006).
Vogel, J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 113, 550–559 (2020).
Acknowledgements
We thank the Synthetic Biology group at Institute Pasteur for discussions. F.B. was supported by the SNSF (P500PB_210944). D.B. and B.B. were supported by the European Research Council (101044479) and Agence Nationale de la Recherche (ANR-10-LABX-62-IBEID). In some instances, we have used ChatGPT to reformulate sentences and improve style. All generated text has been carefully reviewed for accuracy.
Author information
Authors and Affiliations
Contributions
F.B. and D.B. conceptualized the project. B.B. and R.L. contributed to structuring the manuscript. F.B., B.B., R.L., A.M., X.D., A.D. and D.B. wrote the manuscript. F.B., B.B. and R.L. prepared figures and tables. D.B. supervised the project. All authors made a substantial contribution to the discussion of content and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The following authors have links to Eligo Bioscience, of which work is cited and discussed in this article: D.B is a co-founder, shareholder and advisor; X.D. is a co-founder, CEO and shareholder; and A.D. is an employee and shareholder. These authors are also listed as inventors on several patents in the field. This relationship has been disclosed to the journal, and all authors declare that there are no other competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Sheng Yang, who co-reviewed with Siqi Yang, and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Benz, F., Beamud, B., Laurenceau, R. et al. CRISPR–Cas therapies targeting bacteria. Nat Rev Bioeng 3, 627–644 (2025). https://doi.org/10.1038/s44222-025-00311-8
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44222-025-00311-8