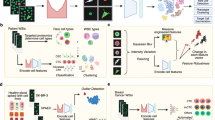
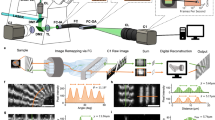
Abstract
The field of genomic research has undergone a remarkable transformation with the rapid expansion of high-quality genome databases, the discovery of epigenetic factors and the advent of CRISPR–Cas technology. However, a major challenge remains in linking these genetic and epigenetic perturbations to spatially resolved cellular phenotypes at scale. Image-activated cell sorting (IACS) offers a way to address this challenge by enabling real-time image-based sorting of suspended objects (for example, single live cells, cell clusters or cells adhered to carriers) at high rates of over 1,000 events per second. Unlike fluorescence-activated cell sorting (FACS), which relies on one-dimensional fluorescence intensity profiles of cells, IACS leverages multi-dimensional optical imaging to capture the full complexity of cellular characteristics, enabling high-content sorting based on both visual and functional attributes. A distinctive feature of IACS is its ability to integrate artificial intelligence for real-time image analysis, enabling sophisticated and precision decision-making in the sorting process. In this Review, we explore the principles, components and key performance indicators of IACS. We also highlight its unique applications across fields such as microbiology, immunology, cancer biology, food science and sustainability science, while addressing the challenges and future opportunities that lie ahead for IACS. Our goal is for this Review to serve as a comprehensive guide to IACS for beginners and experienced users alike, fostering its adoption and driving discoveries across biology and medicine.
Key points
-
The field of genomic research has been revolutionized by the rapid growth of high-quality genome databases, the identification of epigenetic factors and the introduction of CRISPR–Cas technology.
-
A key challenge remains in linking genetic changes to spatially resolved cellular phenotypes, given that spatial organization is vital to understanding cellular function and behaviour.
-
Image-activated cell sorting (IACS) offers a way to address this challenge by providing real-time, high-content sorting of suspended objects such as single live cells, cell clusters or cells adhered to carriers, based on their visual and functional attributes using multi-dimensional imaging.
-
The integration of artificial intelligence for real-time image analysis enables sophisticated and precise decision-making based on nuanced features during the sorting process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Gibbs, R. A. The human genome project changed everything. Nat. Rev. Genet. 21, 575–576 (2020).
Logsdon, G. A., Vollger, M. R. & Eichler, E. E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597–614 (2020).
Altshuler, D. L. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Risch, N. J. Searching for genetic determinants in the new millennium. Nature 405, 847–856 (2000).
Gosden, R. G. & Feinberg, A. P. Genetics and epigenetics — nature’s pen-and-pencil set. N. Engl. J. Med. 356, 731–733 (2007).
Hinte, L. C. et al. Adipose tissue retains an epigenetic memory of obesity after weight loss. Nature 636, 457–465 (2024).
Tomar, A. et al. Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs. Nature 630, 720–727 (2024).
Anway, M. D., Cupp, A. S., Uzumcu, M. & Skinner, M. K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308, 1466–1469 (2005).
Feinberg, A. P. Epigenetics at the epicenter of modern medicine. JAMA 299, 1345–1350 (2008).
Weaver, I. C. G. et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Fraga, M. F. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA 102, 10604–10609 (2005).
Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8, 286–298 (2007).
Karlić, R., Chung, H.-R., Lasserre, J., Vlahoviček, K. & Vingron, M. Histone modification levels are predictive for gene expression. Proc. Natl Acad. Sci. USA 107, 2926–2931 (2010).
Barrangou, R. & Doudna, J. A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Shivram, H., Cress, B. F., Knott, G. J. & Doudna, J. A. Controlling and enhancing CRISPR systems. Nat. Chem. Biol. 17, 10–19 (2021).
Knott, G. J. & Doudna, J. A. CRISPR–Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Barrangou, R. & Horvath, P. A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2, 17092 (2017).
Katti, A., Diaz, B. J., Caragine, C. M., Sanjana, N. E. & Dow, L. E. CRISPR in cancer biology and therapy. Nat. Rev. Cancer 22, 259–279 (2022).
Jordão, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019).
Kotliar, D. et al. Single-cell profiling of Ebola virus disease in vivo reveals viral and host dynamics. Cell 183, 1383–1401.e19 (2020).
Fiskin, E. et al. Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC. Nat. Biotechnol. 40, 374–381 (2022).
Valdes, A. M., Glass, D. & Spector, T. D. Omics technologies and the study of human ageing. Nat. Rev. Genet. 14, 601–607 (2013).
Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018).
Zhang, S. et al. Inference of cell type-specific gene regulatory networks on cell lineages from single cell omic datasets. Nat. Commun. 14, 3064 (2023).
Nolan, T. M. et al. Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science 379, eadf4721 (2023).
Lee, T. I. et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804 (2002).
Caicedo, J. C. et al. Data-analysis strategies for image-based cell profiling. Nat. Methods 14, 849–863 (2017).
Boutros, M., Heigwer, F. & Laufer, C. Microscopy-based high-content screening. Cell 163, 1314–1325 (2015).
Hao, N., Budnik, B. A., Gunawardena, J. & O’Shea, E. K. Tunable signal processing through modular control of transcription factor translocation. Science 339, 460–464 (2013).
Mackinder, L. C. M. et al. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147.e14 (2017).
Moor, A. E. et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 357, 1299–1303 (2017).
Pernas, L., Bean, C., Boothroyd, J. C. & Scorrano, L. Mitochondria restrict growth of the intracellular parasite Toxoplasma gondii by limiting its uptake of fatty acids. Cell Metab. 27, 886–897.e4 (2018).
von Erlach, T. C. et al. Cell-geometry-dependent changes in plasma membrane order direct stem cell signalling and fate. Nat. Mater. 17, 237–242 (2018).
Zenker, J. et al. A microtubule-organizing center directing intracellular transport in the early mouse embryo. Science 357, 925–928 (2017).
He, F. et al. π-HuB: the proteomic navigator of the human body. Nature 636, 322–331 (2024).
Levsky, J. M., Shenoy, S. M., Pezo, R. C. & Singer, R. H. Single-cell gene expression profiling. Science 297, 836–840 (2002).
Edsgärd, D., Johnsson, P. & Sandberg, R. Identification of spatial expression trends in single-cell gene expression data. Nat. Methods 15, 339–342 (2018).
Ludwig, C. H. & Bintu, L. Mapping chromatin modifications at the single cell level. Development 146, dev170217 (2019).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001.e19 (2021).
Albert, F. W., Treusch, S., Shockley, A. H., Bloom, J. S. & Kruglyak, L. Genetics of single-cell protein abundance variation in large yeast populations. Nature 506, 494–497 (2014).
Wu, M. & Singh, A. K. Single-cell protein analysis. Curr. Opin. Biotechnol. 23, 83–88 (2012).
Nitta, N. et al. Intelligent image-activated cell sorting. Cell 175, 266–276.e13 (2018). This pioneering study established IACS as a real-time, AI-driven platform for high-content cell sorting based on intracellular and intercellular phenotypes.
Nitta, N. et al. Raman image-activated cell sorting. Nat. Commun. 11, 3452 (2020). This work presents Raman IACS for chemical imaging-based cell sorting without fluorescent labelling, useful for IACS applications where labelling should be avoided.
Isozaki, A. et al. Intelligent image-activated cell sorting 2.0. Lab. Chip 20, 2263–2273 (2020). This work presents the next-generation intelligent IACS platform with 20-fold improvements in throughput and sensitivity, enabling real-time, high-content sorting of live cells for advanced applications across biology and medicine.
Nawaz, A. A. et al. Intelligent image-based deformation-assisted cell sorting with molecular specificity. Nat. Methods 17, 595–599 (2020). This study introduces a cell-sorting platform to achieve label-free, image-based sorting with molecular specificity by combining deformability cytometry, standing surface acoustic wave actuation and deep learning.
Schraivogel, D. et al. High-speed fluorescence image–enabled cell sorting. Science 375, 315–320 (2022). This study presents an IACS system that integrates fluorescence imaging with droplet sorting to enable genome-scale, phenotype-driven CRISPR screens at unprecedented throughput.
Filby, A. & Carpenter, A. E. A new image for cell sorting. N. Engl. J. Med. 386, 1755–1758 (2022).
Kuhn, T. M., Paulsen, M. & Cuylen-Haering, S. Accessible high-speed image-activated cell sorting. Trends Cell Biol. 34, 657–670 (2024).
Doan, M. & Carpenter, A. E. Leveraging machine vision in cell-based diagnostics to do more with less. Nat. Mater. 18, 414–418 (2019).
LaBelle, C. A., Massaro, A., Cortés-Llanos, B., Sims, C. E. & Allbritton, N. L. Image-based live cell sorting. Trends Biotechnol. 39, 613–623 (2021).
Schraivogel, D. & Steinmetz, L. M. Cell sorters see things more clearly now. Mol. Syst. Biol. 19, e11254 (2023).
Chen, X. et al. Image-guided cell sorting using fast scanning lasers. APL Photon. 5, 040801 (2020).
Herbig, M. et al. Label-free imaging flow cytometry for analysis and sorting of enzymatically dissociated tissues. Sci. Rep. 12, 963 (2022).
Salek, M. et al. COSMOS: a platform for real-time morphology-based, label-free cell sorting using deep learning. Commun. Biol. 6, 971 (2023).
Lee, J. et al. Versatile phenotype-activated cell sorting. Sci. Adv. 6, eabb7438 (2020).
Hasle, N. et al. High‐throughput, microscope‐based sorting to dissect cellular heterogeneity. Mol. Syst. Biol. 16, e9442 (2020).
Nawaz, A. A. et al. Image-based cell sorting using focused travelling surface acoustic waves. Lab. Chip 23, 372–387 (2023).
Lee, K., Kim, S.-E., Doh, J., Kim, K. & Chung, W. K. User-friendly image-activated microfluidic cell sorting technique using an optimized, fast deep learning algorithm. Lab. Chip 21, 1798–1810 (2021).
Gu, Y. et al. Machine learning based real‐time image‐guided cell sorting and classification. Cytometry A 95, 499–509 (2019). This work introduces a real-time, machine learning-based IACS system that combines high-throughput flow cytometry with high-content image analysis and on-chip piezoelectric actuation to enable precise, phenotype-driven cell classification and sorting.
Sesen, M. & Whyte, G. Image-based single cell sorting automation in droplet microfluidics. Sci. Rep. 10, 8736 (2020).
Anagnostidis, V. et al. Deep learning guided image-based droplet sorting for on-demand selection and analysis of single cells and 3D cell cultures. Lab. Chip 20, 889–900 (2020).
Gerling, T., Godino, N., Pfisterer, F., Hupf, N. & Kirschbaum, M. High-precision, low-complexity, high-resolution microscopy-based cell sorting. Lab. Chip 23, 3172–3185 (2023).
Li, Y. et al. Deep cytometry: deep learning with real-time inference in cell sorting and flow cytometry. Sci. Rep. 9, 11088 (2019).
Isozaki, A. et al. A practical guide to intelligent image-activated cell sorting. Nat. Protoc. 14, 2370–2415 (2019). This practical protocol for building and operating an IACS system enabled real-time, image-based sorting using deep learning and an integrated high-throughput microfluidic-optical system.
Tang, R. et al. Low-latency label-free image-activated cell sorting using fast deep learning and AI inferencing. Biosens. Bioelectron. 220, 114865 (2023).
Hayashi, M. et al. Is AI essential? Examining the need for deep learning in image-activated sorting of Saccharomyces cerevisiae. Lab. Chip 23, 4232–4244 (2023).
Harmon, J. et al. Intelligent image‐activated sorting of Chlamydomonas reinhardtii by mitochondrial localization. Cytometry A 101, 1027–1034 (2022).
Isozaki, A. et al. AI on a chip. Lab. Chip 20, 3074–3090 (2020).
Cossarizza, A. et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 49, 1457–1973 (2019).
Cossarizza, A. et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur. J. Immunol. 51, 2708–3145 (2021).
Zhao, Y. et al. Intelligent sort‐timing prediction for image‐activated cell sorting. Cytometry A 103, 88–97 (2023).
Jiao, Z., Zhao, J., Chao, Z., You, Z. & Zhao, J. An air-chamber-based microfluidic stabilizer for attenuating syringe-pump-induced fluctuations. Microfluid. Nanofluid 23, 26 (2019).
Yun, H., Kim, K. & Lee, W. G. Cell manipulation in microfluidics. Biofabrication 5, 022001 (2013).
Patel, Y. M. et al. An inexpensive microfluidic device for three-dimensional hydrodynamic focusing in imaging flow cytometry. Biomicrofluidics 14, 064110 (2020).
Yan, S. & Yuan, D. Continuous microfluidic 3D focusing enabling microflow cytometry for single-cell analysis. Talanta 221, 121401 (2021).
Zhang, P., Bachman, H., Ozcelik, A. & Huang, T. J. Acoustic microfluidics. Annu. Rev. Anal. Chem. 13, 17–43 (2020).
Li, Y. et al. Recent advances in acoustic microfluidics and its exemplary applications. Biomicrofluidics 16, 031502 (2022).
Di Carlo, D. Inertial microfluidics. Lab. Chip 9, 3038 (2009).
Xiang, N. & Ni, Z. Inertial microfluidics: current status, challenges, and future opportunities. Lab. Chip 22, 4792–4804 (2022).
Zhang, T. et al. Passive microfluidic devices for cell separation. Biotechnol. Adv. 71, 108317 (2024).
Lee, K. C. M. et al. Dispersion-free inertial focusing (DIF) for high-yield polydisperse micro-particle filtration and analysis. Lab. Chip 24, 4182–4197 (2024).
Lee, K. C. M., Guck, J., Goda, K. & Tsia, K. K. Toward deep biophysical cytometry: prospects and challenges. Trends Biotechnol. 39, 1249–1262 (2021).
Rees, P., Summers, H. D., Filby, A., Carpenter, A. E. & Doan, M. Imaging flow cytometry. Nat. Rev. Meth. Prim. 2, 86 (2022).
Mikami, H. et al. Virtual-freezing fluorescence imaging flow cytometry. Nat. Commun. 11, 1162 (2020). This optomechanical fluorescence imaging method achieved microscopy-grade imaging at >10,000 eps, overcoming the trade-off between speed, resolution and sensitivity in flow cytometry, an important requirement for IACS.
Doan, M. et al. Deepometry, a framework for applying supervised and weakly supervised deep learning to imaging cytometry. Nat. Protoc. 16, 3572–3595 (2021).
Mikami, H. et al. Ultrafast confocal fluorescence microscopy beyond the fluorescence lifetime limit. Optica 5, 117 (2018).
Kanno, H., Mikami, H., Kaya, Y., Ozeki, Y. & Goda, K. Simple, stable, compact implementation of frequency-division-multiplexed microscopy by inline interferometry. Opt. Lett. 44, 467 (2019).
Diebold, E. D., Buckley, B. W., Gossett, D. R. & Jalali, B. Digitally synthesized beat frequency multiplexing for sub-millisecond fluorescence microscopy. Nat. Photon. 7, 806–810 (2013).
Kanno, H. et al. High-throughput fluorescence lifetime imaging flow cytometry. Nat. Commun. 15, 7376 (2024).
Han, Y. & Lo, Y.-H. Imaging cells in flow cytometer using spatial-temporal transformation. Sci. Rep. 5, 13267 (2015).
Han, Y. et al. Cameraless high-throughput three-dimensional imaging flow cytometry. Optica 6, 1297 (2019).
Lai, Q. T. K. et al. High-speed laser-scanning biological microscopy using FACED. Nat. Protoc. 16, 4227–4264 (2021).
Wang, M. et al. Low-latency in situ image analytics with FPGA-based quantized convolutional neural network. IEEE Trans. Neural Netw. Learn. Syst. 33, 2853–2866 (2022).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Moen, E. et al. Deep learning for cellular image analysis. Nat. Meth. 16, 1233–1246 (2019).
Goda, K., Lu, H., Fei, P. & Guck, J. Revolutionizing microfluidics with artificial intelligence: a new dawn for lab-on-a-chip technologies. Lab. Chip 23, 3737–3740 (2023).
Abadi, M. et al. TensorFlow: a system for large-scale machine learning. In Proc. 12th USENIX Symp. Operating Systems Design and Implementation (OSDI ’16) 265–283 (USENIX, 2016).
Chollet, F. Keras: the Python deep learning library. Keras https://keras.io (2015).
Sakuma, S., Kasai, Y., Hayakawa, T. & Arai, F. On-chip cell sorting by high-speed local-flow control using dual membrane pumps. Lab. Chip 17, 2760–2767 (2017).
Wyatt Shields, C. IV, Reyes, C. D. & López, G. P. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab. Chip 15, 1230–1249 (2015).
Shen, Y., Yalikun, Y. & Tanaka, Y. Recent advances in microfluidic cell sorting systems. Sens. Actuat. B 282, 268–281 (2019).
Wu, M. et al. Acoustofluidic separation of cells and particles. Microsyst. Nanoeng. 5, 32 (2019).
Kim, C., Hwang, D. H., Lee, S. & Kim, S.-J. Water-head pumps provide precise and fast microfluidic pumping and switching versus syringe pumps. Microfluid. Nanofluid. 20, 4 (2016).
Guo, B. & Jiang, L. Flowing cells stability test and evaluation for fast flow cytometry. J. Opt. 48, 54–59 (2019).
Herbig, M. et al. Best practices for reporting throughput in biomedical research. Nat. Meth. 19, 633–634 (2022). This work establishes the best practices for defining and reporting throughput in biomedical research, providing a rigorous framework essential for evaluating high-throughput technologies such as IACS.
Ryan, K., Rose, R. E., Jones, D. R. & Lopez, P. A. Sheath fluid impacts the depletion of cellular metabolites in cells afflicted by sorting induced cellular stress (SICS). Cytometry A 99, 921–929 (2021).
Gala de Pablo, J., Lindley, M., Hiramatsu, K. & Goda, K. High-throughput Raman flow cytometry and beyond. Acc. Chem. Res. 54, 2132–2143 (2021).
Nishiyama, R. et al. Fourier transform coherent anti-Stokes Raman scattering spectroscopy: a comprehensive review. Anal. Chem. 96, 18322–18336 (2024).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Edn 57, 4143–4148 (2018).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Xu, M. et al. Morphological indicator for directed evolution of Euglena gracilis with a high heavy metal removal efficiency. Environ. Sci. Technol. 55, 7880–7889 (2021).
Lei, C. et al. High-throughput optical imaging technology for large-scale single-cell analysis of live Euglena gracilis. TrAC Trends Anal. Chem. 180, 117938 (2024).
Fu, F. & Wang, Q. Removal of heavy metal ions from wastewaters: a review. J. Environ. Manag. 92, 407–418 (2011).
Mehta, S. K. & Gaur, J. P. Use of algae for removing heavy metal ions from wastewater: progress and prospects. Crit. Rev. Biotechnol. 25, 113–152 (2005).
Leong, Y. K. & Chang, J.-S. Bioremediation of heavy metals using microalgae: recent advances and mechanisms. Bioresour. Technol. 303, 122886 (2020).
Lee, K. S. et al. An automated Raman-based platform for the sorting of live cells by functional properties. Nat. Microbiol. 4, 1035–1048 (2019).
Lee, K. S. et al. Optofluidic Raman-activated cell sorting for targeted genome retrieval or cultivation of microbial cells with specific functions. Nat. Protoc. 16, 634–676 (2021).
Stepanauskas, R. et al. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat. Commun. 8, 84 (2017).
Müller, S. & Nebe-von-Caron, G. Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 34, 554–587 (2010).
Lo, M. C. K. et al. Information‐distilled generative label‐free morphological profiling encodes cellular heterogeneity. Adv. Sci. 11, e2307591 (2024).
Siu, D. M. D. et al. Optofluidic imaging meets deep learning: from merging to emerging. Lab. Chip 23, 1011–1033 (2023).
de Rutte, J. et al. Suspendable hydrogel nanovials for massively parallel single-cell functional analysis and sorting. ACS Nano 16, 7242–7257 (2022).
Udani, S. et al. Associating growth factor secretions and transcriptomes of single cells in nanovials using SEC-seq. Nat. Nanotechnol. 19, 354–363 (2024).
Koo, D. et al. Optimizing cell therapy by sorting cells with high extracellular vesicle secretion. Nat. Commun. 15, 4870 (2024).
Koo, D. et al. Defining T cell receptor repertoires using nanovial-based binding and functional screening. Proc. Natl Acad. Sci. 121, e2320442121 (2024).
Zeng, H. & Sanes, J. R. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546 (2017).
Rees, C. L., Moradi, K. & Ascoli, G. A. Weighing the evidence in Peters’ rule: does neuronal morphology predict connectivity? Trends Neurosci. 40, 63–71 (2017).
Martin, D., Xu, J., Porretta, C. & Nichols, C. D. Neurocytometry: flow cytometric sorting of specific neuronal populations from human and rodent brain. ACS Chem. Neurosci. 8, 356–367 (2017).
Spitzer, D. et al. A flow cytometry-based protocol for syngenic isolation of neurovascular unit cells from mouse and human tissues. Nat. Protoc. 18, 1510–1542 (2023).
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Meth. Prim. 2, 8 (2022).
Findinier, J. et al. Dramatic changes in mitochondrial subcellular location and morphology accompany activation of the CO2 concentrating mechanism. Proc. Natl Acad. Sci. 121, e2407548121 (2024).
Isozaki, A. et al. Investigating T‐cell receptor dynamics under in vitro antibody‐based stimulation using imaging flow cytometry. Cytometry A 107, 88–97 (2025).
Challa, D. et al. Function-first discovery of high affinity monoclonal antibodies using Nanovial-based plasma B cell screening. Preprint at bioRxiv https://doi.org/10.1101/2024.08.15.608174 (2024).
Langerman, J. et al. Linking single-cell transcriptomes with secretion using SEC-seq. Nat. Protoc. 20, 2034–2055 (2025).
Vickovic, S. et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 13, 795 (2022).
Kleiber, A., Kraus, D., Henkel, T. & Fritzsche, W. Review: tomographic imaging flow cytometry. Lab. Chip 21, 3655–3666 (2021).
Gualda, E. J., Pereira, H., Martins, G. G., Gardner, R. & Moreno, N. Three‐dimensional imaging flow cytometry through light‐sheet fluorescence microscopy. Cytometry A 91, 144–151 (2017).
Zhang, Z. et al. A high-throughput technique to map cell images to cell positions using a 3D imaging flow cytometer. Proc. Natl. Acad. Sci. 119, e2118068119 (2022).
Cheng, J.-X. & Xie, X. S. Coherent anti-stokes Raman scattering microscopy: instrumentation, theory, and applications. J. Phys. Chem. B 108, 827–840 (2004).
Lindley, M. et al. High‐throughput Raman‐activated cell sorting in the fingerprint region. Adv. Mater. Technol. 7, 2101567 (2022).
Suzuki, Y. et al. Label-free chemical imaging flow cytometry by high-speed multicolor stimulated Raman scattering. Proc. Natl Acad. Sci. 116, 15842–15848 (2019).
Wakisaka, Y. et al. Probing the metabolic heterogeneity of live Euglena gracilis with stimulated Raman scattering microscopy. Nat. Microbiol. 1, 16124 (2016).
Saar, B. G. et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 330, 1368–1370 (2010).
Brzozowski, K. et al. Stimulated Raman scattering microscopy in chemistry and life science — development, innovation, perspectives. Biotechnol. Adv. 60, 108003 (2022).
Vicidomini, G., Bianchini, P. & Diaspro, A. STED super-resolved microscopy. Nat. Meth. 15, 173–182 (2018).
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780 (1994).
Hell, S. W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Lelek, M. et al. Single-molecule localization microscopy. Nat. Rev. Meth. Prim. 1, 39 (2021).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Meth. 3, 793–796 (2006).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Heintzmann, R. & Huser, T. Super-resolution structured illumination microscopy. Chem. Rev. 117, 13890–13908 (2017).
Wu, Y. & Shroff, H. Faster, sharper, and deeper: structured illumination microscopy for biological imaging. Nat. Meth. 15, 1011–1019 (2018).
Li, X. et al. Three-dimensional structured illumination microscopy with enhanced axial resolution. Nat. Biotechnol. 41, 1307–1319 (2023).
Schermelleh, L. et al. Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84 (2019).
Moerner, W. E. & William, E. Single‐molecule spectroscopy, imaging, and photocontrol: foundations for super‐resolution microscopy. Angew. Chem. Int. Edn 54, 8067–8093 (2015).
Keshava, N. & Mustard, J. F. Spectral unmixing. IEEE Signal. Process. Mag. 19, 44–57 (2002).
Nolan, J. P. & Condello, D. Spectral flow cytometry. Curr. Protoc. Cytom. https://doi.org/10.1002/0471142956.cy0127s63 (2013).
Martino, N. et al. Wavelength-encoded laser particles for massively multiplexed cell tagging. Nat. Photon. 13, 720–727 (2019).
Kwok, S. J. J. et al. High-dimensional multi-pass flow cytometry via spectrally encoded cellular barcoding. Nat. Biomed. Eng. 8, 310–324 (2023).
Lugli, E., Roederer, M. & Sottile, R. Multipass high-dimensional flow cytometry. Nat. Biomed. Eng. 8, 209–211 (2023).
Bröker, F., Holt, L. L., Roads, B. D., Dayan, P. & Love, B. C. Demystifying unsupervised learning: how it helps and hurts. Trends Cogn. Sci. 28, 974–986 (2024).
Wu, Z. et al. DynaMorph: self-supervised learning of morphodynamic states of live cells. Mol. Biol. Cell 33, ar59 (2022).
Zhou, Y. et al. Intelligent classification of platelet aggregates by agonist type. eLife 9, e52938 (2020).
Zaritsky, A. et al. Interpretable deep learning uncovers cellular properties in label-free live cell images that are predictive of highly metastatic melanoma. Cell Syst. 12, 733–747.e6 (2021).
Kobayashi, H., Cheveralls, K. C., Leonetti, M. D. & Royer, L. A. Self-supervised deep learning encodes high-resolution features of protein subcellular localization. Nat. Meth. 19, 995–1003 (2022).
Dwivedi, R. et al. Explainable AI (XAI): core ideas, techniques, and solutions. ACM Comput. Surv. 55, 194 (2023).
Novakovsky, G., Dexter, N., Libbrecht, M. W., Wasserman, W. W. & Mostafavi, S. Obtaining genetics insights from deep learning via explainable artificial intelligence. Nat. Rev. Genet. 24, 125–137 (2023).
Ng, D. P. et al. Recommendations for using artificial intelligence in clinical flow cytometry. Cytometry B 106, 228–238 (2024).
Vyawahare, S. et al. Sorting droplets into many outlets. Lab. Chip 21, 4262–4273 (2021).
Sobahi, N. & Han, A. High-throughput and label-free multi-outlet cell counting using a single pair of impedance electrodes. Biosens. Bioelectron. 166, 112458 (2020).
Verbist, W. et al. SeParate: multiway fluorescence-activated droplet sorting based on integration of serial and parallel triaging concepts. Lab. Chip 24, 2107–2121 (2024).
Chen, Y. et al. Pulsed laser activated cell sorting with three dimensional sheathless inertial focusing. Small 10, 1746–1751 (2014).
Iino, T. et al. High-speed microparticle isolation unlimited by Poisson statistics. Lab. Chip 19, 2669–2677 (2019).
Pritchard, R. H. et al. Cell sorting actuated by a microfluidic inertial vortex. Lab. Chip 19, 2456–2465 (2019).
Hu, T., Chitnis, N., Monos, D. & Dinh, A. Next-generation sequencing technologies: an overview. Hum. Immunol. 82, 801–811 (2021).
De Coster, W., Weissensteiner, M. H. & Sedlazeck, F. J. Towards population-scale long-read sequencing. Nat. Rev. Genet. 22, 572–587 (2021).
Jia, Q., Chu, H., Jin, Z., Long, H. & Zhu, B. High-throughput single-сell sequencing in cancer research. Signal. Transduct. Target. Ther. 7, 145 (2022).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016).
Neto, M. D., Oliveira, M. B. & Mano, J. F. Microparticles in contact with cells: from carriers to multifunctional tissue modulators. Trends Biotechnol. 37, 1011–1028 (2019).
Zheng, Y. et al. Microfluidic engineering of crater–terrain hydrogel microparticles: toward novel cell carriers. ACS Appl. Mater. Interf. 15, 7833–7840 (2023).
Zheng, Q. et al. Hydrogels as carriers deliver stem cells/exosomes for liver injury. Mater. Adv. 5, 3587–3601 (2024).
Feng, Q. et al. Dynamic and cell-infiltratable hydrogels as injectable carrier of therapeutic cells and drugs for treating challenging bone defects. ACS Cent. Sci. 5, 440–450 (2019).
Zhuang, S., Semenec, L., Nagy, S. S., Cain, A. K. & Inglis, D. W. High‐precision screening and sorting of double emulsion droplets. Cytometry A 105, 547–554 (2024).
Brower, K. K. et al. Double emulsion flow cytometry with high-throughput single droplet isolation and nucleic acid recovery. Lab. Chip 20, 2062–2074 (2020).
Hulett, H. R., Bonner, W. A., Barrett, J. & Herzenberg, L. A. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science 166, 747–749 (1969).
Liu, L., Cheung, T. H., Charville, G. W. & Rando, T. A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 10, 1612–1624 (2015).
Przybyla, L. & Gilbert, L. A. A new era in functional genomics screens. Nat. Rev. Genet. 23, 89–103 (2022).
Shapiro, H. M. Practical Flow Cytometry (John Wiley & Sons, 2005).
Cho, S. H., Chen, C. H., Tsai, F. S., Godin, J. M. & Lo, Y.-H. Human mammalian cell sorting using a highly integrated micro-fabricated fluorescence-activated cell sorter (μFACS). Lab. Chip 10, 1567 (2010).
Chen, C. H. et al. Specific sorting of single bacterial cells with microfabricated fluorescence-activated cell sorting and tyramide signal amplification fluorescence in situ hybridization. Anal. Chem. 83, 7269–7275 (2011).
Acknowledgements
This work was supported by the JSPS Core-to-Core Program (JPJSCCA20190007 to K.G.), the Research Grants Council (14125924 and RFS2021-7S06 to K.K.T.), the Innovation and Technology Commission (InnoHK initiative and ITS/318/22FP to K.K.T.), the Chan Zuckerberg Initiative Donor Advised Fund (2023-332386 to D.D.C.), the Humboldt Foundation’s Philipp Franz von Siebold Award (to K.G.), the Humboldt Association of Japan (to K.G.), the Mohammed bin Salman Center for Future Science and Technology for Saudi-Japan Vision 2030 (to K.G.), and the Cell Aging Control Endowed Chair at the University of Tokyo. We gratefully acknowledge the Serendipity Lab, the MSCA ITN Cell2Cell, and the LMU Munich and the University of Tokyo strategic partnership fund for facilitating collaboration opportunities.
Author information
Authors and Affiliations
Contributions
All authors contributed to the writing of the manuscript. K.G. edited the manuscript and led the manuscript development team.
Corresponding author
Ethics declarations
Competing interests
K.G. is a shareholder of CYBO, LucasLand and FlyWorks. K.K.T. is a shareholder of Conzeb. D.D.C. and the Regents of the University of California are shareholders of Partillion Bioscience Corporation, which sells Nanovials.
Citation diversity statement
We acknowledge that papers authored by scholars from minoritized groups are systematically under-cited. Here, we have made every attempt to reference relevant papers in a manner that is equitable in terms of racial, ethnic, gender and geographical representation.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Yu-Hwa Lo, Shi-Wei Chu, who co-reviewed with Hsuan-Wen Lin, and Paul Rees for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ding, T., Lee, K.C.M., Tsia, K.K. et al. Image-activated cell sorting. Nat Rev Bioeng 3, 890–907 (2025). https://doi.org/10.1038/s44222-025-00334-1
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44222-025-00334-1
This article is cited by
-
Label-free blood cell separation for space health monitoring using a portable blast cell biochip
npj Microgravity (2026)
-
Single-pixel imaging flow cytometry for biomedical research
Inflammation and Regeneration (2025)