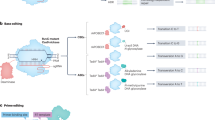
Abstract
CRISPR genome-editing tools, including Cas9 and Cas12a nucleases, base editors and prime editors, have revolutionized genome manipulation across various species and cell types. These tools have undergone continuous improvement as new variants or types of editors have been generated to improve their efficiency, specificity and applicability. However, given the vast array of genome editors and the multitude of designable guide RNAs, selecting the optimal combinations for efficient and precise genome editing has become increasingly challenging, especially under variable experimental conditions. To address this issue, several methods for evaluating genome-editing tools in a high-throughput manner have been developed. The resulting large datasets of editing efficiencies or specificities have been used to develop machine learning models that predict efficiency and specificity, greatly facilitating the optimal selection of genome editors and guide RNAs. Here, we review recent developments in high-throughput evaluations and machine learning-based predictions of genome-editing efficiencies and/or off-target effects, together with recent advances in diverse genome-editing tools. We also cover artificial intelligence-based development and evolution of genome-editing tools.
Key points
-
Gene-editing tools such as CRISPR nucleases, base editors and prime editors can manipulate genomes across a wide range of species and cell types.
-
With the expansion of the genome-editing toolbox, the selection of the most suitable tool and the design of appropriate guide RNAs for precise and efficient editing remain challenging.
-
High-throughput methods enable the measurement of protospacer adjacent motif sequence preference, on-target and off-target editing efficiencies, and outcomes at diverse target sequences, facilitating the determination of optimal genome-editing tools and guide RNA combinations from the available options.
-
The large amount of data on genome-editing tools evaluated at thousands of target DNA sequences has facilitated the development of machine learning models that quickly predict genome-editing activity for a given pair of guide RNA and target sequence.
-
Large language models trained on DNA, RNA and protein data enable the artificial intelligence-based design and evolution of genome editors. Combined with high-throughput datasets, they can contribute to the development of next-generation genome-editing tools.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
DiCarlo, J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013).
Jiang, W. Y., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D. G. & Kamoun, S. Targeted mutagenesis in the model plant using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693 (2013).
Shan, Q. W. et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688 (2013).
Wang, H. Y. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Zou, Q. J. et al. Generation of gene-target dogs using CRISPR/Cas9 system. J. Mol. Cell Biol. 7, 580–583 (2015).
Hai, T., Teng, F., Guo, R. F., Li, W. & Zhou, Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24, 372–375 (2014).
Niu, Y. Y. et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843 (2014).
Koike-Yusa, H., Li, Y. L., Tan, E. P., Velasco-Herrera, M. D. & Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 (2014).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Hart, T. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015).
Hanna, R. E. et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e20 (2021).
Kim, Y. et al. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nat. Biotechnol. 40, 874–884 (2022).
Sánchez-Rivera, F. J. et al. Base editing sensor libraries for high-throughput engineering and functional analysis of cancer-associated single nucleotide variants. Nat. Biotechnol. 40, 862–873 (2022).
Martin-Rufino, J. D. et al. Massively parallel base editing to map variant effects in human hematopoiesis. Cell 186, 2456–2474.e24 (2023).
Erwood, S. et al. Saturation variant interpretation using CRISPR prime editing. Nat. Biotechnol. 40, 885–895 (2022).
Ren, X. J. et al. High-throughput PRIME-editing screens identify functional DNA variants in the human genome. Mol. Cell 83, 4633–4645.e9 (2023).
Gould, S. I. et al. High-throughput evaluation of genetic variants with prime editing sensor libraries. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02172-9 (2024).
Findlay, G. M. et al. Accurate classification of variants with saturation genome editing. Nature 562, 217–222 (2018).
Frangoul, H. et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 384, 252–260 (2021).
Cetin, B., Erendor, F., Eksi, Y. E., Sanlioglu, A. D. & Sanlioglu, S. Advancing CRISPR genome editing into gene therapy clinical trials: progress and future prospects. Expert Rev. Mol. Med. 27, e16 (2025).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Cox, D. B. T. et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017).
Esvelt, K. M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121 (2013).
Harrington, L. B. et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 (2018).
Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021).
Kim, D. Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40, 94–102 (2022).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
Urnov, F. D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005).
Chen, F. Q. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Rouet, P., Smih, F. & Jasin, M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14, 8096–8106 (1994).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–542 (2015).
Riesenberg, S. et al. Efficient high-precision homology-directed repair-dependent genome editing by HDRobust. Nat. Methods 20, 1388–1399 (2023).
Panier, S. & Boulton, S. J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18 (2014).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Kleinstiver, B. P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Chen, J. S. et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Casini, A. et al. A highly specific SpCas9 variant is identified by screening in yeast. Nat. Biotechnol. 36, 265–271 (2018).
Lee, J. K. et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 9, 3048 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018).
Miller, S. M. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 38, 471–481 (2020).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020).
Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021).
Xu, X. et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell 81, 4333–4345.e4 (2021).
Han, D. et al. Development of miniature base editors using engineered IscB nickase. Nat. Methods 20, 1029–1036 (2023).
Han, L. et al. Engineering miniature IscB nickase for robust base editing with broad targeting range. Nat. Chem. Biol. 20, 1629–1639 (2024).
Xue, N. et al. Engineering IscB to develop highly efficient miniature editing tools in mammalian cells and embryos. Mol. Cell 84, 3128–3140.e4 (2024).
Hino, T. et al. An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis. Cell 186, 4920–4935.e23 (2023).
Li, Z. et al. Engineering a transposon-associated TnpB-ωRNA system for efficient gene editing and phenotypic correction of a tyrosinaemia mouse model. Nat. Commun. 15, 831 (2024).
Yan, H. et al. Assessing and engineering the IscB–ωRNA system for programmed genome editing. Nat. Chem. Biol. 20, 1617–1628 (2024).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Smargon, A. A. et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018).
Özcan, A. et al. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 597, 720–725 (2021).
van Beljouw, S. P. B. et al. The gRAMP CRISPR-Cas effector is an RNA endonuclease complexed with a caspase-like peptidase. Science 373, 1349–1353 (2021).
Abbott, T. R. et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 181, 865–876.e12 (2020).
Blanchard, E. L. et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. 39, 717–726 (2021).
Liang, W. W. et al. Transcriptome-scale RNA-targeting CRISPR screens reveal essential lncRNAs in human cells. Cell 187, 7637–7654.e29 (2024).
Montero, J. J. et al. Genome-scale pan-cancer interrogation of lncRNA dependencies using CasRx. Nat. Methods 21, 584–596 (2024).
Wang, L. et al. CRISPR-Cas13d screens identify KILR, a breast cancer risk-associated lncRNA that regulates DNA replication and repair. Mol. Cancer 23, 101 (2024).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Joung, J. et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 383, 1492–1494 (2020).
Tong, H. et al. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat. Biotechnol. 41, 108–119 (2023).
Zhang, F. et al. Multiplexed inhibition of immunosuppressive genes with Cas13d for combinatorial cancer immunotherapy. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02535-2 (2025).
Hwang, G. H. et al. Large DNA deletions occur during DNA repair at 20-fold lower frequency for base editors and prime editors than for Cas9 nucleases. Nat. Biomed. Eng. 9, 79–92 (2025).
Wu, L. et al. Adenine base editors induce off-target structure variations in mouse embryos and primary human T cells. Genome Biol. 25, 291 (2024).
Grünewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Yu, Y. et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 11, 2052 (2020).
Lam, D. K. et al. Improved cytosine base editors generated from TadA variants. Nat. Biotechnol. 41, 686–697 (2023).
Neugebauer, M. E. et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol. 41, 673–685 (2023).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Kannan, S. et al. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 40, 194–197 (2022).
Li, G. et al. Developing PspCas13b-based enhanced RESCUE system, eRESCUE, with efficient RNA base editing. Cell Commun. Signal. 19, 84 (2021).
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021).
Doman, J. L. et al. Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell 186, 3983–4002.e26 (2023).
Yan, J. et al. Improving prime editing with an endogenous small RNA-binding protein. Nature 628, 639–647 (2024).
Jang, H. et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 6, 181–194 (2022).
Liu, B. et al. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat. Biotechnol. 40, 1388–1393 (2022).
She, K. et al. Dual-AAV split prime editor corrects the mutation and phenotype in mice with inherited retinal degeneration. Signal Transduct. Target. Ther. 8, 57 (2023).
Davis, J. R. et al. Efficient prime editing in mouse brain, liver and heart with dual AAVs. Nat. Biotechnol. 42, 253–264 (2024).
Nelson, J. W. et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410 (2022).
Kwon, J. et al. TAPE-seq is a cell-based method for predicting genome-wide off-target effects of prime editor. Nat. Commun. 13, 7975 (2022).
Liang, S. Q. et al. Genome-wide profiling of prime editor off-target sites in vitro and in vivo using PE-tag. Nat. Methods 20, 898–907 (2023).
Kim, D., Moon, S. B., Ko, J. H., Kim, Y. S. & Kim, D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 48, 10576–10589 (2020).
Karvelis, T. et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 253 (2015).
Walton, R. T., Hsu, J. Y., Joung, J. K. & Kleinstiver, B. P. Scalable characterization of the PAM requirements of CRISPR–Cas enzymes using HT-PAMDA. Nat. Protoc. 16, 1511–1547 (2021).
Doench, J. G. et al. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Guo, J. H. et al. Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Res. 46, 7052–7069 (2018).
Kleinstiver, B. P. et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298 (2015).
Chari, R., Mali, P., Moosburner, M. & Church, G. M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods 12, 823–826 (2015).
Kim, H. K. et al. In vivo high-throughput profiling of CRISPR–Cpf1 activity. Nat. Methods 14, 153–159 (2017).
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018).
Allen, F. et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 37, 64–72 (2019).
Kim, H. K. et al. SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci. Adv. 5, eaax9249 (2019).
Kim, N. et al. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 38, 1328–1336 (2020).
Wang, D. Q. et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 10, 4284 (2019).
Kim, H. K. et al. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat. Biomed. Eng. 4, 111–124 (2020).
Seo, S. Y. et al. Massively parallel evaluation and computational prediction of the activities and specificities of 17 small Cas9s. Nat. Methods 20, 999–1009 (2023).
Arbab, M. et al. Determinants of base editing outcomes from target library analysis and machine learning. Cell 182, 463–480.e30 (2020).
Song, M. et al. Sequence-specific prediction of the efficiencies of adenine and cytosine base editors. Nat. Biotechnol. 38, 1037–1043 (2020).
Marquart, K. F. et al. Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens. Nat. Commun. 12, 5114 (2021).
Zhang, C. D. et al. Prediction of base editor off-targets by deep learning. Nat. Commun. 14, 5358 (2023).
Kim, N. et al. Deep learning models to predict the editing efficiencies and outcomes of diverse base editors. Nat. Biotechnol. 42, 484–497 (2024).
Pallaseni, A. et al. Predicting base editing outcomes using position-specific sequence determinants. Nucleic Acids Res. 50, 3551–3564 (2022).
Yu, G. et al. Prediction of efficiencies for diverse prime editing systems in multiple cell types. Cell 186, 2256–2272.e23 (2023).
Kim, H. K. et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 39, 198–206 (2021).
Koeppel, J. et al. Prediction of prime editing insertion efficiencies using sequence features and DNA repair determinants. Nat. Biotechnol. 41, 1446–1456 (2023).
Mathis, N. et al. Predicting prime editing efficiency and product purity by deep learning. Nat. Biotechnol. 41, 1151–1159 (2023).
Mathis, N. et al. Machine learning prediction of prime editing efficiency across diverse chromatin contexts. Nat. Biotechnol. 43, 712–719 (2025).
Akhtar, W. et al. Using TRIP for genome-wide position effect analysis in cultured cells. Nat. Protoc. 9, 1255–1281 (2014).
Kim, Y., Oh, H. C., Lee, S. & Kim, H. H. Saturation profiling of drug-resistant genetic variants using prime editing. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02465-z (2024).
Sack, L. M., Davoli, T., Xu, Q. K., Li, M. M. Z. & Elledge, S. J. Sources of error in mammalian genetic screens. G3 Genes Genomes Genet. 6, 2781–2790 (2016).
Schlub, T. E., Smyth, R. P., Grimm, A. J., Mak, J. & Davenport, M. P. Accurately measuring recombination between closely related HIV-1 genomes. PLoS Comput. Biol. 6, e1000766 (2010).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Wienert, B. et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289 (2019).
Lazzarotto, C. R. et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol. 38, 1317–1327 (2020).
Kim, D. & Kim, J. S. DIG-seq: a genome-wide CRISPR off-target profiling method using chromatin DNA. Genome Res. 28, 1894–1900 (2018).
Zou, R. S. et al. Improving the sensitivity of in vivo CRISPR off-target detection with DISCOVER-Seq+. Nat. Methods 20, 706–713 (2023).
Kim, D., Kim, D. E., Lee, G., Cho, S. I. & Kim, J. S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 37, 430–435 (2019).
Liang, P. et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 10, 67 (2019).
Zhu, M. et al. Tracking-seq reveals the heterogeneity of off-target effects in CRISPR–Cas9-mediated genome editing. Nat. Biotechnol. 43, 799–810 (2025).
Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015).
Wong, N., Liu, W. J. & Wang, X. W. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218 (2015).
Xu, H. et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157 (2015).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Kim, H. K. et al. Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat. Biotechnol. 36, 239–241 (2018).
Xiang, X. et al. Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun. 12, 3238 (2021).
Wessels, H. H. et al. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 38, 722–727 (2020).
Wessels, H. H. et al. Prediction of on-target and off-target activity of CRISPR–Cas13d guide RNAs using deep learning. Nat. Biotechnol. 42, 628–637 (2024).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016).
Listgarten, J. et al. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng. 2, 38–47 (2018).
Zhang, S. X., Li, X. T., Lin, Q. Z. & Wong, K. C. Synergizing CRISPR/Cas9 off-target predictions for ensemble insights and practical applications. Bioinformatics 35, 1108–1115 (2019).
Jiang, K. et al. Rapid in silico directed evolution by a protein language model with EVOLVEpro. Science 387, eadr6006 (2024).
Nguyen, E. et al. Sequence modeling and design from molecular to genome scale with Evo. Science 386, eado9336 (2024).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Ryu, S. M. et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539 (2018).
Villiger, L. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525 (2018).
Koblan, L. W. et al. In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature 589, 608–614 (2021).
Davis, J. R. et al. Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat. Biomed. Eng. 6, 1272–1283 (2022).
Böck, D. et al. In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 14, eabl9238 (2022).
Lin, Q. P. et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927 (2021).
Anzalone, A. V. et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731–740 (2022).
Choi, J. H. et al. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 40, 218–226 (2022).
Wang, J. L. et al. Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 19, 331–340 (2022).
Zhuang, Y. et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat. Chem. Biol. 18, 29–37 (2022).
Yarnall, M. T. N. et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol. 41, 500–512 (2023).
Pandey, S. et al. Efficient site-specific integration of large genes in mammalian cells via continuously evolved recombinases and prime editing. Nat. Biomed. Eng. 9, 22–39 (2025).
Klein, J. C. et al. Multiplex pairwise assembly of array-derived DNA oligonucleotides. Nucleic Acids Res. 44, e43 (2016).
Hegde, M., Strand, C., Hanna, R. E. & Doench, J. G. Uncoupling of sgRNAs from their associated barcodes during PCR amplification of combinatorial CRISPR screens. PLoS ONE 13, e0197547 (2018).
Williams, R. et al. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 3, 545–550 (2006).
Feldman, D., Singh, A., Garrity, A. J. & Blainey, P. C. Lentiviral co-packaging mitigates the effects of intermolecular recombination and multiple integrations in pooled genetic screens. Preprint at bioRxiv https://doi.org/10.1101/262121 (2018).
Behan, F. M. et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516 (2019).
Lawson, K. A. et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 586, 120–126 (2020).
Marceau, C. D. et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163 (2016).
Daniloski, Z. et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell 184, 92–105.e16 (2021).
Dong, M. B. et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204.e23 (2019).
Acknowledgements
This work was supported, in part, by the National Research Foundation of Korea grant funded by the Korean government (Ministry of Science and ICT) (RS-2024-00348839 (H.K.K.), RS-2022-NR070713 (H.H.K.), and RS-2025-02214844 (H.H.K.)), The Bio and Medical Technology Development Program of the National Research Foundation funded by the Korean government (Ministry of Science and ICT) (RS-2023-00262533 (H.K.K.), RS-2022-NR067326 (H.H.K.), RS-2022-NR067345 (H.H.K.) and RS-2023-00260968 (H.H.K.)), the Korea Drug Development Fund funded by the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy (RS-2022-DD128884 (H.K.K.)), the Korea–US Collaborative Research Fund (KUCRF) funded by the Ministry of Science and ICT and Ministry of Health & Welfare, Republic of Korea (grant number: RS-2024-00467177 (H.H.K.)), the Yonsei Signature Research Cluster Program of 2024-22-0165 (H.H.K.), the Brain Korea 21 FOUR Project for Medical Science (Yonsei University College of Medicine), the SNUH Kun-hee Lee Child Cancer and Rare Disease Project, Republic of Korea (22B-000-0101 (H.H.K.)), and the Yonsei Fellow Program, funded by Lee Youn Jae.
Author information
Authors and Affiliations
Contributions
H.H.K. conceptualized the manuscript. H.K.K. and H.H.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.H.K. is the founder of cisionMed. H.K.K. declares no competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Yongming Wang, Sidi Chen, who co-reviewed with Kaiyuan Tang and Lei Peng, and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, H.K., Kim, H.H. Evaluation and prediction of guide RNA activities in genome-editing tools. Nat Rev Bioeng 4, 82–97 (2026). https://doi.org/10.1038/s44222-025-00352-z
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44222-025-00352-z