Abstract
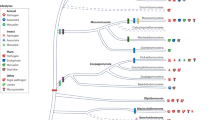
The class Agaricomycetes represents the mushroom-forming fungi; it is a diverse group within the Basidiomycota with an evolutionary history spanning over 350 million years. Interest in Agaricomycetes has grown considerably over the past decades because of their important roles in nature, their developmental biology, and their use in green technologies and medicine. In this Review, we discuss how genomics approaches have contributed to important breakthroughs in understanding their ecology, evolution, development and conservation. We also explore the central challenges constraining further research. We postulate that the surge in omics exploration of Agaricomycetes over the past decade will be followed by a postgenomic era combining experimental, reverse genetics, ecological and functional genomics tools, helping to resolve the recalcitrant questions surrounding the complex biology of these fungi.
Key points
-
The class Agaricomycetes is a taxonomically and functionally diverse group of fungi with >350 million years of evolutionary history.
-
They are among the most important recyclers of dead plant biomass and are mutualistic partners of woody plants.
-
A robust phylogenetic framework allows reconstruction of ancestral morphologies and lifestyles with high confidence.
-
Agaricomycete genomics has advanced tremendously in the past decade. Further progress will require a shift to postgenomic and functional approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lutzoni, F. et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat. Commun. 9, 5451 (2018).
Hibbett, D. S. After the gold rush, or before the flood? Evolutionary morphology of mushroom-forming fungi (Agaricomycetes) in the early 21st century. Mycol. Res. 111, 1001–1018 (2007).
Kiss, E. et al. Comparative genomics reveals the origin of fungal hyphae and multicellularity. Nat. Commun. 10, 4080 (2019).
Money, N. P. Goldilocks mushrooms: how ballistospory has shaped basidiomycete evolution. Fungal Biol. 127, 975–984 (2023).
Money, N. P., Stolze, J. & Fischer, M. W. F. Mechanics of the artillery fungus. Fungal Biol. 128, 2334–2340 (2024).
Hofrichter, M. ed. The MYCOTA X. Industrial Applications (Springer Berlin Heidelberg, 2011).
Baldrian, P. Wood-inhabiting ligninolytic basidiomycetes in soils: ecology and constraints for applicability in bioremediation. Fungal Ecol. 1, 4–12 (2008).
Põlme, S. et al. Fungaltraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 105, 1–16 (2020).
Correction for Riley et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl Acad. Sci. USA 111, 14959–14959 (2014).
Pan, Y. et al. A large and persistent carbon sink in the world’s forests. Science 333, 988–993 (2011).
Floudas, D. Evolution of lignin decomposition systems in fungi. Adv. Botanical Res. 99, 37–76 (2021).
Floudas, D. et al. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719 (2012).
Nelsen, M. P., DiMichele, W. A., Peters, S. E. & Boyce, C. K. Delayed fungal evolution did not cause the paleozoic peak in coal production. Proc. Natl Acad. Sci. USA 113, 2442–2447 (2016).
Eastwood, D. C. et al. The plant cell wall–decomposing machinery underlies the functional diversity of forest fungi. Science 333, 762–765 (2011).
Floudas, D. et al. Evolution of novel wood decay mechanisms in agaricales revealed by the genome sequences of fistulina hepatica and cylindrobasidium torrendii. Fungal Genet. Biol. 76, 78–92 (2015).
Nagy, L. G. et al. Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol. Biol. Evol. 33, 959–970 (2016).
Arantes, V. & Goodell, B. in ACS Symposium Series Vol. 1158 (eds. Schultz, T. P., Goodell, B. & Nicholas, D. D.) 3–21 (American Chemical Society, 2014).
Jensen, K. A., Ryan, Z. C., Vanden Wymelenberg, A., Cullen, D. & Hammel, K. E. An NADH:quinone oxidoreductase active during biodegradation by the brown-rot basidiomycete Gloeophyllum trabeum. Appl. Env. Microbiol. 68, 2699–2703 (2002).
Suzuki, M. R., Hunt, C. G., Houtman, C. J., Dalebroux, Z. D. & Hammel, K. E. Fungal hydroquinones contribute to brown rot of wood. Environ. Microbiol. 8, 2214–2223 (2006).
Korripally, P., Timokhin, V. I., Houtman, C. J., Mozuch, M. D. & Hammel, K. E. Evidence from serpula lacrymans that 2,5-dimethoxyhydroquinone is a lignocellulolytic agent of divergent brown rot basidiomycetes. Appl. Env. Microbiol. 79, 2377–2383 (2013).
Floudas, D. et al. X-ray scattering reveals two mechanisms of cellulose microfibril degradation by filamentous fungi. Appl. Env. Microbiol. 88, e00995-22 (2022).
Goodell, B. et al. Modification of the nanostructure of lignocellulose cell walls via a non-enzymatic lignocellulose deconstruction system in brown rot wood-decay fungi. Biotechnol. Biofuels 10, 179 (2017).
Marlin, M., Wolf, A., Alomran, M., Carta, L. & Newcombe, G. Nematophagous pleurotus species consume some nematode species but are themselves consumed by others. Forests 10, 404 (2019).
Riley, R. et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl Acad. Sci. USA 111, 9923–9928 (2014).
Floudas, D. et al. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 14, 2046–2059 (2020).
Almási, É et al. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytol. 224, 902–915 (2019).
Larsson, K.-H. Re-thinking the classification of corticioid fungi. Mycol. Res. 111, 1040–1063 (2007).
Harder, C. B. et al. Mycena species can be opportunist-generalist plant root invaders. Environ. Microbiol. 25, 1875–1893 (2023).
Morin, E. et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl Acad. Sci. USA 109, 17501–17506 (2012).
Four, B., Cárdenas, R. E. & Dangles, O. Traits or habitat? Disentangling predictors of leaf-litter decomposition in amazonian soils and streams. Ecosphere 10, e02691 (2019).
Ruiz-Dueñas, F. J. et al. Genomic analysis enlightens agaricales lifestyle evolution and increasing peroxidase diversity. Mol. Biol. Evol. 38, 1428–1446 (2021).
Koch, R. A. et al. Marasmioid rhizomorphs in bird nests: Species diversity, functional specificity, and new species from the tropics. Mycologia 112, 1086–1103 (2020).
Schultz, T. R. et al. The coevolution of fungus–ant agriculture. Science 386, 105–110 (2024).
Mueller, U. G. & Gerardo, N. Fungus-farming insects: multiple origins and diverse evolutionary histories. Proc. Natl Acad. Sci. USA 99, 15247–15249 (2002).
Aanen, D. K. et al. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl Acad. Sci. USA 99, 14887–14892 (2002).
Aanen, D. K. & Eggleton, P. Fungus-growing termites originated in african rain forest. Curr. Biol. 15, 851–855 (2005).
Nygaard, S. et al. Reciprocal genomic evolution in the ant–fungus agricultural symbiosis. Nat. Commun. 7, 12233 (2016).
Aguilar-Colorado, ÁS. & Rivera-Chávez, J. Ants/nest-associated fungi and their specialized metabolites: taxonomy, chemistry, and bioactivity. Rev. Bras. Farmacogn. 33, 901–923 (2023).
van de Peppel, L. J. J. et al. Ancestral predisposition toward a domesticated lifestyle in the termite-cultivated fungus termitomyces. Curr. Biol. 31, 4413–4421 (2021).
Anthony, M. A. et al. Forest tree growth is linked to mycorrhizal fungal composition and function across Europe. ISME J. 16, 1327–1336 (2022).
Strullu-Derrien, C., Selosse, M., Kenrick, P. & Martin, F. M. The origin and evolution of mycorrhizal symbioses: from palaeomycology to phylogenomics. New Phytol. 220, 1012–1030 (2018).
Tedersoo, L. & Smith, M. E. Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 27, 83–99 (2013).
Kohler, A. et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47, 410–415 (2015).
Corrales, A. et al. Diversity and distribution of tropical ectomycorrhizal fungi. Mycologia 114, 919–933 (2022).
Medina-Vega, J. A. et al. Tropical tree ectomycorrhiza are distributed independently of soil nutrients. Nat. Ecol. Evol. 8, 400–410 (2024).
Steidinger, B. S. et al. Climatic controls of decomposition drive the global biogeography of forest-tree symbioses. Nature 569, 404–408 (2019).
Tedersoo, L., Bahram, M. & Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 367, eaba1223 (2020).
Smith, S. E. & Read, D. J. Mycorrhizal Symbiosis (Academic Press, 2010).
Koele, N., Dickie, I. A., Oleksyn, J., Richardson, S. J. & Reich, P. B. No globally consistent effect of ectomycorrhizal status on foliar traits. New Phytol. 196, 845–852 (2012).
Bunn, R. A. et al. What determines transfer of carbon from plants to mycorrhizal fungi? New Phytol. 244, 1199–1215 (2024).
Stuart, E. K. et al. Species-level identity of Pisolithus influences soil phosphorus availability for host plants and is moderated by nitrogen status, but not CO2. Soil. Biol. Biochem. 165, 108520 (2022).
Berrios, L. & Peay, K. G. Field reduction of ectomycorrhizal fungi has cascading effects on soil microbial communities and reduces the abundance of ectomycorrhizal symbiotic bacteria. Mol. Ecol. 34, e17585 (2025).
Branco, S., Schauster, A., Liao, H. & Ruytinx, J. Mechanisms of stress tolerance and their effects on the ecology and evolution of mycorrhizal fungi. New Phytol. 235, 2158–2175 (2022).
Colpaert, J. V., Wevers, J. H. L., Krznaric, E. & Adriaensen, K. How metal-tolerant ecotypes of ectomycorrhizal fungi protect plants from heavy metal pollution. Ann. For. Sci. 68, 17–24 (2011).
Bazzicalupo, A. L. et al. Fungal heavy metal adaptation through single nucleotide polymorphisms and copy-number variation. Mol. Ecol. 29, 4157–4169 (2020).
Zhang, K., Tappero, R., Ruytinx, J., Branco, S. & Liao, H.-L. Disentangling the role of ectomycorrhizal fungi in plant nutrient acquisition along a Zn gradient using X-ray imaging. Sci. Total. Environ. 801, 149481 (2021).
Smith, A. et al. Comparative transcriptomics provides insights into molecular mechanisms of zinc tolerance in the ectomycorrhizal fungus Suillus luteus. G3 14, jkae156 (2024).
Pellegrin, C., Morin, E., Martin, F. M. & Veneault-Fourrey, C. Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front. Microbiol. 6, 1278 (2015).
Miyauchi, S. et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat. Commun. 11, 5125 (2020).
Zhang, F. et al. The ectomycorrhizal basidiomycete laccaria bicolor releases a GH28 polygalacturonase that plays a key role in symbiosis establishment. New Phytol. 233, 2534–2547 (2022).
Plett, J. M. et al. Speciation underpinned by unexpected molecular diversity in the mycorrhizal fungal genus pisolithus. Mol. Biol. Evol. 40, msad045 (2023).
Plett, J. M. et al. Effector MiSSP7 of the mutualistic fungus laccaria bicolor stabilizes the populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl Acad. Sci. USA 111, 8299–8304 (2014).
Kang, H. et al. The small secreted effector protein MiSSP7.6 of Laccaria bicolor is required for the establishment of ectomycorrhizal symbiosis. Environ. Microbiol. 22, 1435–1446 (2020).
Wong-Bajracharya, J. et al. The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc. Natl Acad. Sci. USA 119, e2103527119 (2022).
Wu, G. et al. Evolutionary innovations through gain and loss of genes in the ectomycorrhizal Boletales. New Phytol. 233, 1383–1400 (2022).
Looney, B. P. et al. Russulaceae: a new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytol. 218, 54–65 (2018).
Plett, K. L., Wojtalewicz, D., Anderson, I. C. & Plett, J. M. Fungal metabolism and free amino acid content may predict nitrogen transfer to the host plant in the ectomycorrhizal relationship between Pisolithus spp. and Eucalyptus grandis. New Phytol. 242, 1589–1602 (2024).
Tremble, K., Hoffman, J. I. & Dentinger, B. T. M. Contrasting continental patterns of adaptive population divergence in the holarctic ectomycorrhizal fungus Boletus edulis. New Phytol. 237, 295–309 (2023).
Branco, S. et al. Continental-level population differentiation and environmental adaptation in the mushroom Suillus brevipes. Mol. Ecol. 26, 2063–2076 (2017).
Stuart, E. K. & Plett, K. L. Digging deeper: in search of the mechanisms of carbon and nitrogen exchange in ectomycorrhizal symbioses. Front. Plant Sci. 10, 1658 (2019).
Plett, J. M. & Plett, K. L. Leveraging genomics to understand the broader role of fungal small secreted proteins in niche colonization and nutrition. ISME Commun. 2, 49 (2022).
Lofgren, L. A. et al. Comparative genomics reveals dynamic genome evolution in host specialist ectomycorrhizal fungi. New Phytol. 230, 774–792 (2021).
Satish, L. et al. Agrobacterium tumefaciens-mediated genetic transformation of the ect-endomycorrhizal fungus Terfezia boudieri. Genes 11, 1293 (2020).
Plett, K. L. et al. Inorganic nitrogen availability alters eucalyptus grandis receptivity to the ectomycorrhizal fungus Pisolithus albus but not symbiotic nitrogen transfer. New Phytol. 226, 221–231 (2020).
Kemppainen, M., Chowdhury, J., Lundberg-Felten, J. & Pardo, A. Fluorescent protein expression in the ectomycorrhizal fungus Laccaria bicolor: a plasmid toolkit for easy use of fluorescent markers in basidiomycetes. Curr. Genet. 66, 791–811 (2020).
Randewig, D., Marshall, J. D., Nasholm, T. & Jamtgard, S. Combining microdialysis with metabolomics to characterize the in situ composition of dissolved organic compounds in boreal forest soil. Soil Biol. Biochem. 136, 107530 (2019).
Plett, K. L. et al. Novel microdialysis technique reveals a dramatic shift in metabolite secretion during the early stages of the interaction between the ectomycorrhizal fungus Pisolithus microcarpus and its host Eucalyptus grandis. Microorganisms 9, 1817 (2021).
Vishwakarma, K. et al. Pisolithus microcarpus isolates with contrasting abilities to colonise Eucalyptus grandis exhibit significant differences in metabolic signalling. Fungal Biol. 128, 2157–2166 (2024).
Lofgren, L. et al. Suillus: an emerging model for the study of ectomycorrhizal ecology and evolution. New Phytol. 242, 1448–1475 (2024).
Olson, Å et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 194, 1001–1013 (2012).
Simard, S. W. et al. Net transfer of carbon between ectomycorrhizal tree species in the field. Nature 388, 579–582 (1997).
Karst, J., Jones, M. D. & Hoeksema, J. D. Positive citation bias and overinterpreted results lead to misinformation on common mycorrhizal networks in forests. Nat. Ecol. Evol. 7, 501–511 (2023).
Bogar, L. M., Tavasieff, O. S., Raab, T. K. & Peay, K. G. Does resource exchange in ectomycorrhizal symbiosis vary with competitive context and nitrogen addition? New Phytol. 233, 1331–1344 (2022).
Baumgartner, K., Coetzee, M. P. A. & Hoffmeister, D. Secrets of the subterranean pathosystem of Armillaria. Mol. Plant Pathol. 12, 515–534 (2011).
Anderson, J. B. et al. Clonal evolution and genome stability in a 2500-year-old fungal individual. Proc. Biol. Sci. 285, 20182233 (2018).
Aime, M. C. & Phillips-Mora, W. The causal agents of witches’ broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia 97, 1012–1022 (2005).
Sipos, G. et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 1, 1931–1941 (2017).
Parfitt, D., Hunt, J., Dockrell, D., Rogers, H. J. & Boddy, L. Do all trees carry the seeds of their own destruction? PCR reveals numerous wood decay fungi latently present in sapwood of a wide range of angiosperm trees. Fungal Ecol. 3, 338–346 (2010).
Sahu, N. et al. Vertical and horizontal gene transfer shaped plant colonization and biomass degradation in the fungal genus Armillaria. Nat. Microbiol. 8, 1668–1681 (2023).
Vasconcelos, A. A. et al. Adaptive evolution of Moniliophthora PR-1 proteins towards its pathogenic lifestyle. BMC Ecol. Evol. 21, 84 (2021).
Anderson, J. P. et al. Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci. Rep. 7, 10410 (2017).
Reina, R. et al. Genome and secretome of Chondrostereum purpureum correspond to saprotrophic and phytopathogenic life styles. PLoS ONE 14, e0212769 (2019).
Matsumoto, R. et al. Genomic and secretomic analyses of the newly isolated fungus Perenniporia fraxinea SS3 identified CAZymes potentially related to a serious pathogenesis of hardwood trees. Appl. Environ. Microbiol. 89, e0027223 (2023).
Redkar, A., Sabale, M., Zuccaro, A. & Di Pietro, A. Determinants of endophytic and pathogenic lifestyle in root colonizing fungi. Curr. Opin. Plant Biol. 67, 102226 (2022).
Varga, T. et al. Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 3, 668–678 (2019).
Sánchez-Ramírez, S., Etienne, R. S. & Moncalvo, J.-M. High speciation rate at temperate latitudes explains unusual diversity gradients in a clade of ectomycorrhizal fungi. Evolution 69, 2196–2209 (2015).
Abarenkov, K. et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered. Nucleic Acids Res. 52, D791–D797 (2024).
van Galen, L. G. et al. The biogeography and conservation of Earth’s ‘dark’ ectomycorrhizal fungi. Curr. Biol. 35, R563–R574 (2025).
Geml, J., Tulloss, R. E., Laursen, G. A., Sazanova, N. A. & Taylor, D. L. Evidence for strong inter- and intracontinental phylogeographic structure in Amanita muscaria, a wind-dispersed ectomycorrhizal basidiomycete. Mol. Phylogenet. Evol. 48, 694–701 (2008).
Pringle, A. & Vellinga, E. C. Last chance to know? Using literature to explore the biogeography and invasion biology of the death cap mushroom Amanita phalloides (Vaill. ex Fr.:Fr.) Link. Biol. Invasions 8, 1131–1144 (2006).
Blackwell, M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98, 426–438 (2011).
Cowie, R. H., Bouchet, P. & Fontaine, B. The sixth mass extinction: fact, fiction or speculation? Biol. Rev. 97, 640–663 (2022).
Mueller, G. M. et al. What do the first 597 global fungal red list assessments tell us about the threat status of fungi? Diversity 14, 736 (2022).
Gange, A. C., Gange, E. G., Sparks, T. H. & Boddy, L. Rapid and recent changes in fungal fruiting patterns. Science 316, 71 (2007).
Kauserud, H. et al. Warming-induced shift in European mushroom fruiting phenology. Proc. Natl Acad. Sci. USA 109, 14488–14493 (2012).
Pietras, M., Kolanowska, M. & Selosse, M.-A. Quo vadis? Historical distribution and impact of climate change on the worldwide distribution of the australasian fungus Clathrus archeri (Phallales, Basidiomycota). Mycol. Progress 20, 299–311 (2021).
A, V., Mt, B., Dl, L., A, P. & Ma, J. Invasive golden oyster mushrooms are disrupting native fungal communities as they spread throughout North America. Curr. Biol. 35, 3994–4002 (2025).
Dickie, I. A. et al. Towards management of invasive ectomycorrhizal fungi. Biol. Invasions 18, 3383–3395 (2016).
Wang, Y.-W. et al. Invasive californian death caps develop mushrooms unisexually and bisexually. Nat. Commun. 14, 6560 (2023).
Egli, S., Peter, M., Buser, C., Stahel, W. & Ayer, F. Mushroom picking does not impair future harvests — results of a long-term study in Switzerland. Biol. Conserv. 129, 271–276 (2006).
Unit, B. Kunming-montreal global biodiversity framework. Convention on Biological Diversity https://www.cbd.int/gbf (2024).
Taylor, J. W., Turner, E., Townsend, J. P., Dettman, J. R. & Jacobson, D. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos. Trans. R. Soc. Lond. B 361, 1947–1963 (2006).
Peay, K. G., Schubert, M. G., Nguyen, N. H. & Bruns, T. D. Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol. Ecol. 21, 4122–4136 (2012).
Norros, V., Penttilä, R., Suominen, M. & Ovaskainen, O. Dispersal may limit the occurrence of specialist wood decay fungi already at small spatial scales. Oikos 121, 961–974 (2012).
Li, D.-W. Release and dispersal of basidiospores from Amanita muscaria var. alba and their infiltration into a residence. Mycol. Res. 109, 1235–1242 (2005).
Simões, T. R. & Pierce, S. E. Sustained high rates of morphological evolution during the rise of tetrapods. Nat. Ecol. Evol. 5, 1403–1414 (2021).
Berendse, F. & Scheffer, M. The angiosperm radiation revisited, an ecological explanation for Darwin’s ‘abominable mystery’. Ecol. Lett. 12, 865–872 (2009).
Cai, C., Leschen, R. A. B., Hibbett, D. S., Xia, F. & Huang, D. Mycophagous rove beetles highlight diverse mushrooms in the cretaceous. Nat. Commun. 8, 14894 (2017).
Poinar, G. & Buckley, R. Evidence of mycoparasitism and hypermycoparasitism in Early Cretaceous amber. Mycol. Res. 111, 503–506 (2007).
Smith, S. Y., Currah, R. S. & Stockey, R. A. Cretaceous and eocene poroid hymenophores from Vancouver Island, British Columbia. Mycologia 96, 180–186 (2004).
Prasanna, A. N. et al. Model choice, missing data, and taxon sampling impact phylogenomic inference of deep basidiomycota relationships. Syst. Biol. 69, 17–37 (2020).
Hibbett, D. S. & Donoghue, M. J. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst. Biol. 50, 215–242 (2001).
Sánchez-García, M. et al. Fruiting body form, not nutritional mode, is the major driver of diversification in mushroom-forming fungi. Proc. Natl Acad. Sci. USA 117, 32528–32534 (2020).
Iapichino, M., Wang, Y.-W., Gentry, S., Pringle, A. & Seminara, A. A precise relationship among Buller’s drop, ballistospore, and gill morphologies enables maximum packing of spores within gilled mushrooms. Mycologia 113, 300–311 (2021).
Money, N. P. The fastest short jump in nature: progress in understanding the mechanism of ballistospore discharge. Fungal Biol. 127, 835–844 (2023).
Sakes, A. et al. Shooting mechanisms in nature: a systematic review. PLoS ONE 11, e0158277 (2016).
Hou, Z. et al. An evolutionarily ancient transcription factor drives spore morphogenesis in mushroom-forming fungi. Curr. Biol. https://doi.org/10.1016/j.cub.2025.02.025 (2025).
Fritz, J. A., Seminara, A., Roper, M., Pringle, A. & Brenner, M. P. A natural O-ring optimizes the dispersal of fungal spores. J. R. Soc. Interface 10, 20130187 (2013).
Hibbett, D. S. Trends in morphological evolution in homobasidiomycetes inferred using maximum likelihood: a comparison of binary and multistate approaches. Syst. Biol. 53, 889–903 (2004).
Virágh, M. et al. Evolutionary morphogenesis of sexual fruiting bodies in basidiomycota: toward a new evo-devo synthesis. Microbiol. Mol. Biol. Rev. 86, e0001921 (2022).
Varga, T., Földi, C., Bense, V. & Nagy, L. G. Radiation of mushroom-forming fungi correlates with novel modes of protecting sexual fruiting bodies. Fungal Biol. 126, 556–565 (2022).
Stafleu, F. A. Evolution of the higher basidiomycetes. Taxon 20, 616–618 (1971).
Thiers, H. D. The secotioid syndrome. Mycologia 76, 1–8 (1984).
Nagy, L. G. et al. The evolution of defense mechanisms correlate with the explosive diversification of autodigesting Coprinellus mushrooms (Agaricales, Fungi). Syst. Biol. 61, 595–607 (2012).
Tóth, A. et al. Iteratively refined guide trees help improving alignment and phylogenetic inference in the mushroom family bolbitiaceae. PLoS ONE 8, e56143 (2013).
Bodensteiner, P., Binder, M., Moncalvo, J.-M., Agerer, R. & Hibbett, S. D. Phylogenetic relationships of cyphelloid homobasidiomycetes. Mol. Phylogenet. Evol. 33, 501–515 (2004).
Krah, F.-S. et al. European mushroom assemblages are darker in cold climates. Nat. Commun. 10, 2890 (2019).
Kuhar, F., Terzzoli, L., Nouhra, E., Robledo, G. & Mercker, M. Pattern formation features might explain homoplasy: fertile surfaces in higher fungi as an example. Theory Biosci. 141, 1–11 (2022).
Peña, A. et al. A multiomic approach to understand how pleurotus eryngii transforms non-woody lignocellulosic material. J. Fungi 7, 426 (2021).
Kohler, A. & Martin, F. in Molecular Mycorrhizal Symbiosis (ed. Martin, F.) 87–106 (Wiley, 2016).
Wolfe, B. E., Tulloss, R. E. & Pringle, A. The irreversible loss of a decomposition pathway marks the single origin of an ectomycorrhizal symbiosis. PLoS ONE 7, e39597 (2012).
Sheikh, S., Khan, F. K., Bahram, M. & Ryberg, M. Impact of model assumptions on the inference of the evolution of ectomycorrhizal symbiosis in fungi. Sci. Rep. 12, 22043 (2022).
Sato, H. The evolution of ectomycorrhizal symbiosis in the Late Cretaceous is a key driver of explosive diversification in Agaricomycetes. New Phytol. 241, 444–460 (2024).
Nakamori, T. & Suzuki, A. Defensive role of cystidia against Collembola in the basidiomycetes Russula bella and Strobilurus ohshimae. Mycol. Res. 111, 1345–1351 (2007).
Cai, Q. et al. The evolution of ectomycorrhizal symbiosis and host-plant switches are the main drivers for diversification of Amanitaceae (Agaricales, Basidiomycota). BMC Biol. 22, 230 (2024).
Vajda, V. & McLoughlin, S. Fungal proliferation at the Cretaceous–Tertiary boundary. Science 303, 1489 (2004).
Givnish, T. J. Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytol. 207, 297–303 (2015).
Wilson, A. W., Binder, M. & Hibbett, D. S. Effects of gasteroid fruiting body morphology on diversification rates in three independent clades of fungi estimated using binary state speciation and extinction analysis. Evolution 65, 1305–1322 (2011).
Ryberg, M. & Matheny, P. B. Dealing with incomplete taxon sampling and diversification of a large clade of mushroom-forming fungi: diversification of a large mushroom-forming clade. Evolution 65, 1862–1878 (2011).
Sánchez-García, M. & Matheny, P. B. Is the switch to an ectomycorrhizal state an evolutionary key innovation in mushroom-forming fungi? A case study in the Tricholomatineae (Agaricales). Evolution 71, 51–65 (2017).
Ryberg, M. & Matheny, P. B. Asynchronous origins of ectomycorrhizal clades of Agaricales. Proc. R. Soc. B. 279, 2003–2011 (2012).
Wilson, A. W., Hosaka, K. & Mueller, G. M. Evolution of ectomycorrhizas as a driver of diversification and biogeographic patterns in the model mycorrhizal mushroom genus Laccaria. New Phytol. 213, 1862–1873 (2017).
Codjia, J. E. I. et al. Historical biogeography and diversification of ringless Amanita (section Vaginatae) support an African origin and suggest niche conservatism in the Americas. Mol. Phylogenet. Evol. 178, 107644 (2023).
Looney, B. P., Ryberg, M., Hampe, F., Sánchez-García, M. & Matheny, P. B. Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Mol. Ecol. 25, 630–647 (2016).
Geml, J., Laursen, G. A., O’Neill, K., Nusbaum, H. C. & Taylor, D. L. Beringian origins and cryptic speciation events in the fly agaric (Amanita muscaria). Mol. Ecol. 15, 225–239 (2006).
Sánchez-Ramírez, S. et al. In and out of refugia: historical patterns of diversity and demography in the North American Caesar’s mushroom species complex. Mol. Ecol. 24, 5938–5956 (2015).
Hage, H. et al. Gene family expansions and transcriptome signatures uncover fungal adaptations to wood decay. Environ Microbiol 23, 5716–5732 (2021).
Zhao, H. et al. Insights into the ecological diversification of the hymenochaetales based on comparative genomics and phylogenomics with an emphasis on Coltricia. Genome Biol. Evol. 15, evad136 (2023).
Looney, B. et al. Evolutionary transition to the ectomycorrhizal habit in the genomes of a hyperdiverse lineage of mushroom-forming fungi. New Phytol. 233, 2294–2309 (2022).
Harder, C. B. et al. Extreme overall mushroom genome expansion in Mycena s.s. irrespective of plant hosts or substrate specializations. Cell Genom. 4, 100586 (2024).
Dentinger, B. T. M. et al. Tales from the crypt: genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol. J. Linn. Soc. 117, 11–32 (2016).
Andrew, C., Diez, J., James, T. Y. & Kauserud, H. Fungarium specimens: a largely untapped source in global change biology and beyond. Philos. Trans. R. Soc. Lond. B 374, 20170392 (2019).
Kües, U. From two to many: multiple mating types in basidiomycetes. Fungal Biol. Rev. 29, 126–166 (2015).
James, T. Y. Why mushrooms have evolved to be so promiscuous: insights from evolutionary and ecological patterns. Fungal Biol. Rev. 29, 167–178 (2015).
Raper, J. R., Krongelb, G. S. & Baxter, M. G. The number and distribution of incompatibility factors in schizophyllum. Am. Nat. 92, 221–232 (1958).
Peris, D. et al. Large-scale fungal strain sequencing unravels the molecular diversity in mating loci maintained by long-term balancing selection. PLoS Genet. 18, e1010097 (2022).
Martin, F. et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452, 88–92 (2008).
Hiltunen, M., Ament-Velásquez, S. L., Ryberg, M. & Johannesson, H. Stage-specific transposon activity in the life cycle of the fairy-ring mushroom Marasmius oreades. Proc. Natl Acad. Sci. USA 119, e2208575119 (2022).
Plett, J. M. et al. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 21, 1197–1203 (2011).
Feldman, D., Kowbel, D. J., Glass, N. L., Yarden, O. & Hadar, Y. A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: ligninolytic enzyme regulation in Pleurotus ostreatus. Sci. Rep. 7, 14553 (2017).
Nagy, L. G., Kovács, G. M. & Krizsán, K. Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biol. Rev. Camb. Philos. Soc. 93, 1778–1794 (2018).
Krizsán, K. et al. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc. Natl Acad. Sci. USA 116, 7409–7418 (2019).
Nagy, L. G. et al. Lessons on fruiting body morphogenesis from genomes and transcriptomes of Agaricomycetes. Stud. Mycol. 104, 1–85 (2023).
Anderson, J. B. & Catona, S. Genomewide mutation dynamic within a long-lived individual of Armillaria gallica. Mycologia 106, 642–648 (2014).
Hiltunen, M., Grudzinska-Sterno, M., Wallerman, O., Ryberg, M. & Johannesson, H. Maintenance of high genome integrity over vegetative growth in the fairy-ring mushroom Marasmius oreades. Curr. Biol. 29, 2758–2765.e6 (2019).
Aanen, D. K. How a long-lived fungus keeps mutations in check. Science 346, 922–923 (2014).
Thorén, M. H. et al. Early germline sequestration in a basidiomycete fungus. Science 389, 720–723 (2025).
Reynolds, H. T. et al. Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evol. Lett. 2, 88–101 (2018).
Bradshaw, A. J. et al. Phylogenomics of the psychoactive mushroom genus Psilocybe and evolution of the psilocybin biosynthetic gene cluster. Proc. Natl Acad. Sci. USA 121, e2311245121 (2024).
Ke, H.-M. et al. Mycena genomes resolve the evolution of fungal bioluminescence. Proc. Natl Acad. Sci. USA 117, 31267–31277 (2020).
Liu, F., Wang, S.-H., Cheewangkoon, R. & Zhao, R.-L. Uneven distribution of prokaryote-derived horizontal gene transfer in fungi: a lifestyle-dependent phenomenon. mBio 16, e02855–24 (2025).
Mudbhari, S. et al. Decoding the chemical language of Suillus fungi: genome mining and untargeted metabolomics uncover terpene chemical diversity. mSystems 9, e0122523 (2024).
Drott, M. T. et al. Pangenomics of the death cap mushroom amanita phalloides, and of agaricales, reveals dynamic evolution of toxin genes in an invasive range. ISME J. 17, 1236–1246 (2023).
Haikazian, S. et al. Psilocybin-assisted therapy for depression: a systematic review and meta-analysis. Psychiatry Res. 329, 115531 (2023).
Luo, H. et al. Genes and evolutionary fates of the amanitin biosynthesis pathway in poisonous mushrooms. Proc. Natl Acad. Sci. USA 119, e2201113119 (2022).
Nofiani, R. et al. Strobilurin biosynthesis in basidiomycete fungi. Nat. Commun. 9, 3940 (2018).
Sum, W. C., Ebada, S. S., Clement Matasyoh, J. & Stadler, M. Recent progress in the evaluation of secondary metabolites from Basidiomycota. Curr. Res. Biotechnol. 6, 100155 (2023).
Keller, N. P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180 (2019).
Kotlobay, A. A. et al. Genetically encodable bioluminescent system from fungi. Proc. Natl Acad. Sci. USA 115, 12728–12732 (2018).
Thoen, E. et al. In vitro evidence of root colonization suggests ecological versatility in the genus Mycena. New Phytol. 227, 601–612 (2020).
Wu, X. et al. The diversity and co-occurrence network of soil bacterial and fungal communities and their implications for a new indicator of grassland degradation. Ecol. Indic. 129, 107989 (2021).
Bödeker, I. T. M. et al. Ectomycorrhizal Cortinarius species participate in enzymatic oxidation of humus in northern forest ecosystems. New Phytol. 203, 245–256 (2014).
Lindahl, B. D. & Tunlid, A. Ectomycorrhizal fungi — potential organic matter decomposers, yet not saprotrophs. New Phytol. 205, 1443–1447 (2015).
Botnen, S. et al. Low host specificity of root-associated fungi at an Arctic site. Mol. Ecol. 23, 975–985 (2014).
Rodriguez, R. J., White, J. F., Arnold, A. E. & Redman, R. S. Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330 (2009).
Almario, J. et al. Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl Acad. Sci. USA 114, E9403–E9412 (2017).
Almario, J., Fabiańska, I., Saridis, G. & Bucher, M. Unearthing the plant-microbe quid pro quo in root associations with beneficial fungi. New Phytol. 234, 1967–1976 (2022).
Yuan, Z. et al. Genomic landscape of a relict fir-associated fungus reveals rapid convergent adaptation towards endophytism. ISME J. 16, 1294–1305 (2022).
Merényi, Z. et al. Gene age shapes the transcriptional landscape of sexual morphogenesis in mushroom-forming fungi (Agaricomycetes). eLife 11, e71348 (2022).
Gehrmann, T. et al. Nucleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regulatory programs. Proc. Natl Acad. Sci. USA 115, 4429–4434 (2018).
Hegedüs, B. et al. Morphogenesis, starvation, and light responses in a mushroom-forming fungus revealed by long-read sequencing and extensive expression profiling. Cell Genom. 5, 100853 (2025).
Wu, B. et al. Retraction note: evolution of substrate-specific gene expression and RNA editing in brown rot wood-decaying fungi. ISME J. 16, 322 (2022).
Peng, L. et al. A facultative ectomycorrhizal association is triggered by organic nitrogen. Curr. Biol. 32, 5235–5249.e7 (2022).
Niedźwiedzki, G., Szrek, P., Narkiewicz, K., Narkiewicz, M. & Ahlberg, P. E. Tetrapod trackways from the early middle devonian period of Poland. Nature 463, 43–48 (2010).
Jiao, Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011).
COL. Catalogue of Life. catalogueoflife.org https://www.catalogueoflife.org/ (2025).
Nilsson, R. H. et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264 (2019).
Robert, V. et al. MycoBank gearing up for new horizons. IMA Fungus 4, 371–379 (2013).
Hughes, A. C. et al. Sampling biases shape our view of the natural world. Ecography 44, 1259–1269 (2021).
Titley, M. A., Snaddon, J. L. & Turner, E. C. Scientific research on animal biodiversity is systematically biased towards vertebrates and temperate regions. PLoS ONE 12, e0189577 (2017).
Větrovský, T. et al. GlobalFungi, a global database of fungal occurrences from high-throughput-sequencing metabarcoding studies. Sci. Data 7, 228 (2020).
GBIF.Org User. Occurrence download. The Global Biodiversity Information Facility https://doi.org/10.15468/DL.AR9XPF (2025).
Hibbett, D. S. A phylogenetic overview of the agaricomycotina. Mycologia 98, 917–925 (2006).
Liu, S.-L., Wei, H.-W. & Zhou, L.-W. Xenasmatellales ord. nov. and Xenasmatellaceae fam. nov. for Xenasmatella (Agaricomycetes, Basidiomycota). Mycology 14, 175–189 (2023).
Berbee, M. L., Wong, E. Y. Y. & Tsui, C. K. M. Phylogenetic evidence places the coralloid jelly fungus Tremellodendropsis tuberosa (Tremellodendropsidales) among early diverging agaricomycetes. Mycol. Prog. 15, 939–946 (2016).
He, M.-Q. et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 99, 105–367 (2019).
Vizzini, A., Alvarado, P., Consiglio, G., Marchetti, M. & Xu, J. Family matters inside the order Agaricales: systematic reorganization and classification of incertae sedis clitocyboid, pleurotoid and tricholomatoid taxa based on an updated 6-gene phylogeny. Stud. Mycol. 107, 67–148 (2024).
Kirk, P. M. et al. Ainsworth and Bisbys Dictionary of the Fungi 10th edn (CABI Publishing, 2008).
Acknowledgements
GBIF, UNITE and MycoBank are acknowledged for assistance with occurrence and bibliographic data for species descriptions. L.G.N. was supported by the European Research Council (grant no. 101086900) and the National Research Development and Innovation Office (grant no. OTKA 142188). S.B. is supported by NSF IOS-PBI 2029168. Z.M. was supported by the Janos Bolyai Research Scholarship of the Hungarian Academy of Sciences (grant no. BO/00269/24/8). F.M.M. is funded by the National Key Laboratory of Ecological Security and Sustainable Development in the Arid Region, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, China (grant no. 23YFFA0013).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article. Z.M and L.G.N. researched data for the article. L.G.N. contributed the vision and coordinated the writing of the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
A.P. is vice president of Mushroom Observer, a US non-profit dedicated to cataloguing fungal mushroom biodiversity. All other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Biodiversity thanks Christoffer Bugge Harder, Håvard Kauserud, Dabao Lu and Martin Rybergand for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
MycoBank: https://www.mycobank.org/
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nagy, L.G., Branco, S., Floudas, D. et al. The biodiversity, genomics, ecology and evolution of mushroom-forming fungi. Nat. Rev. Biodivers. 2, 24–39 (2026). https://doi.org/10.1038/s44358-025-00107-z
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44358-025-00107-z