Abstract
Immunohistochemical analyses (IHC) of biomarkers are extensively used for tumor characterization and as prognostic and predictive measures. The current standard of single slide analysis assumes that one 5 μM section is representative of the entire tumor. We used our automated image analysis technology (AQUA) using a modified IHC technique with fluorophores to compare estrogen receptor (ER) expression in multiple blocks/slides from cases of primary breast cancer with the objective of quantifying tumor heterogeneity within sections and between blocks. To normalize our ER scores and allow slide-to-slide comparisons, 0.6 μm histospots of representative breast cancer cases with known ER scores were assembled into a ‘gold standard array’ (GSA) and placed adjacently to each whole section. Overall, there was excellent correlation between AQUA scores and the pathologist's scores and reproducibility of GSA scores (mean linear regression R value 0.8903). Twenty-nine slides from 11 surgical cases were then analyzed totaling over 2000 AQUA images. Using standard binary assignments of AQUA (>10) and pathologist's (>10%) scores as being positive, there was fair concordancy between AQUA and pathologist scores (73%) and between slides from different blocks from the same cases (75%). However using continuous AQUA scores, agreement between AQUA and pathologist was far lower and between slides from different blocks from the same cases only 19%. Within individual slides there was also significant heterogeneity in a scattered pattern, most notably for slides with the highest AQUA scores. In sum, using a quantitative measure of ER expression, significant block-to-block heterogeneity was found in 81% of cases. These results most likely reflect both laboratory-based variability due to lack of standardization of immunohistochemistry and true biological heterogeneity. It is also likely to be dependent on the biomarker analyzed and suggests further studies should be carried out to determine how these findings may affect clinical decision-making processes.
Similar content being viewed by others
Main
It has long been recognized that breast cancer exhibits widely disparate clinical behavior that cannot be solely attributable to stage. Recent expression profiling studies have corroborated this finding by identifying biologically distinct groups of tumors spanning traditional classification schemas such as stage and tumor size.1 Within an individual tumor, biologically relevant heterogeneity may also exist in part, owing to variations in the tumor microenvironment, cell cycle variations, and stem cell subpopulations.
Immunohistochemical (IHC) evaluation of biomarkers in tissue has evolved to become a commonly used diagnostic tool for the pathologist, predominantly because of its ease and ability to retain morphologic information. In addition, tissue microarrays (TMA) have become a popular tool for the rapid and efficient detection of clinicopathologic associations in large numbers of samples.2, 3 Major criticisms of TMAs relate to tumor heterogeneity and whether small TMA cores are representative of the whole section. On a broader scale, these criticisms are limited because they are based on the assumption that whole sections accurately reflect the entire tumor. For example, tissue is typically sampled at the rate of one section per cm3 of tumor. Although the volume of each TMA spot represents only about 0.0002% (0.6 × 0.6 mm2 diameter and 5 μm thick) of this tumor, the standard tissue section would represent only 0.05% (1 × 1 cm dimension and 5 μm thick) of this tumor.
Estrogen receptor (ER) is an important regulator of both physiologic and pathologic mammary growth and differentiation.4, 5 Although its expression has been associated with well differentiated, lower grade tumors, most studies have shown that it also retains independent prognostic information.6 More importantly, ER expression is the most reliable predictor of response to endocrine therapies in breast cancer.7 Current standard of care for determining ER status of a given clinical sample is IHC on a whole section with manual readouts. Although individual tumors typically have relatively homogenous ER expression, staining heterogeneity has been observed in some tumors.8
We have developed an algorithm for quantitatively determining in situ protein expression called automated image analysis technology (AQUA).9 AQUA is a hybrid of standard IHC and flow cytometry in that it requires antigen retrieval on fixed tissue, application of primary and secondary antibodies, and use of multiplexed fluorescent detection to produce an objective, numeric score. This technology reduces the bias of subjective assessment and allows quantification of protein expression using molecular colocalization techniques. Unlike either technology however, the resultant score is directly proportional to the concentration of the target protein within a user-defined compartment.10 This methodology has been validated with a variety of biomarkers in many different cancers.11, 12, 13 Thus, we used AQUA to collect continuous ER scores from multiple images/fields of standard whole-tissue sections from multiple blocks of the same surgical cases with the objective of quantifying tumor heterogeneity.
Materials and methods
Case Selection
Eleven cases of primary invasive ductal adenocarcinomas of the breast from 1999 to 2000 were retrieved from the Yale University Pathology archives. Specifically, cases were selected that spanned a broad range of ER expression as determined by the pathologist at routine signout (eg 0–100%), that had abundance of tumor present in the blocks, and that had multiple blocks available from the same surgical case. Areas of invasive tumor were identified by a pathologist and circled on the whole section, giving careful attention to avoid areas with admixed in situ and/or benign tissue. Overall, there were 29 slides from 11 cases (either one, two, or three blocks from the same case and one section studied per each block). This study was approved by the Yale University Human Investigations Committee.
Gold Standard Array/Control Array
In order to normalize our ER scores and allow slide-to-slide comparisons, a ‘gold standard array’ (GSA) of exemplary, representative breast cancer cases with known ER scores was constructed. Six cases of breast cancer from 2002 were selected from the archives of the Yale University Department of Pathology. These cases also had abundant invasive cancer in the blocks and had a relatively homogenous appearance to the tumor. The signout pathologist's ER scores were 0, 20, 50, 70, 80, and 100% and the pathologists in this study felt these were classic or exemplary examples of these scores. Representative areas of invasive tumor were identified by a pathologist and 0.6 mm diameter cores were placed into a recipient block using a precision arraying instrument (Beecher Instruments, Silver Spring, MD, USA). First, a master/triplicate GSA was assembled by taking three consecutive cuts (5 μm) of the GSA recipient block and affixing to an adhesive slide using a UV crosslinkable tape transfer system. Then, one 5 μm cut of the GSA was affixed onto slides adjacent to each whole section from the 29 slides described above to be stained and analyzed concurrently.
Immunohistochemistry
Staining slides for AQUA has been previously described.9 Briefly, slides were deparaffinized in xylene, rinsed in ethanol, and rehydrated. Antigen retrieval was performed by pressure cooking for 15 min in 6.5 mM sodium citrate buffer. Endogenous peroxidase was quenched by immersing the array in a 2.5% methanol/hydrogen peroxide buffer for 30 min. Nonspecific background staining was further minimized by preincubating the array with 0.3% bovine serum albumin in 0.1 M Tris-buffered saline (pH 8.0) for 1 h. Primary antibodies used were pancytokeratin and ER, clone 1D5 (DAKO, Carpinteria, CA, USA). This primary antibody cocktail was incubated overnight at 4°C in a humidity chamber. Goat anti-mouse antibody conjugated to a horseradish peroxidase-decorated dextran polymer backbone (Envision; DAKO Corp.) was used as a secondary reagent to detect the bound primary target (ER) and Cy5-tyramide was used to visualize the amplified signal. Cy-5 (red) was used because its emission peak is well outside the green-orange spectrum of tissue autofluorescence. The cytokeratin was visualized with a Cy3-conjugated secondary antibody and the array was then counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to define the nuclear compartment.
Image Collection and AQUA Analysis
Image acquisition and automated analysis have also been described extensively in previous work.9 For the whole section analysis, images for the GSA and whole sections on each slide were captured separately. Images are automatically acquired with a high-resolution monochromatic camera using filter cubes specific to the emission/excitation spectra of DAPI, Cy5, and Cy3. Then, using this stack of uncompressed images, the AQUA software then allows one to distinguish between areas of tumor and stromal elements using the cytokeratin stain, resulting in a unique binary cytokeratin tumor mask for each spot. Furthermore, the cytokeratin and DAPI stains are used to assign each pixel under the tumor mask into non-overlapping membrane/cytoplasmic and nuclear locales. AQUA scores for ER are then calculated that correspond to the average signal intensity divided by compartment area. Although non-nuclear ER expression may be biologically relevant, as standard ER analysis relies on nuclear expression, we chose to quantitate ER signal in the nuclear compartment, that is, AQUA ER score within the DAPI-defined nuclear compartment. The AQUA score is thus proportional to the average protein concentration in the nucleus across all of the cells within the keratin staining mask. This information can then be exported in a format suitable for analysis by standard statistical software packages.
For the whole sections, using a × 20 objective, a series of image ‘fields’ were captured within the circled invasive tumor to ultimately cover the vast majority of tissue of interest. Depending on the size of the tumor, 33–147 fields were captured per section and a total of 2043 fields were analyzed. Postimage capture, images were reviewed, particularly near the edges, to ensure that only fields within circled area of invasive tumor were included for analysis.
Results
Analysis of GSA and Normalization
Three consecutive 5 μm cuts of the GSA were placed on the same slide, stained with cytokeratin, ER, and DAPI and analyzed with AQUA. Figure 1a shows the mean of the master/triplicate AQUA scores compared with the pathologist's ER scores. Overall, there was an excellent correlation between the mean AQUA scores and the pathologist scores. However, because the AQUA scores represent ER concentration, and whereas the pathologist scores represent simply the percent nuclei positive without regard to signal intensity or concentration, the relationship is not strictly linear.
Gold standard array analysis. AQUA ER scores on three-fold redundant TMAs (triplicate GSA) were averaged and compared with the pathologist's ER estimation (a). These scores were then compared with the GSA scores (same cases as those on the triplicate GSA) on each whole-section analyzed to serve as internal controls and as a means of normalizing each whole section raw AQUA scores with the Master/Triplicate GSA.
Next, log AQUA scores of the GSA histospots on each whole-section slide was compared with the mean log scores on the master/triplicate GSA. Of note, identical staining conditions were used for all slides, including antibody concentrations, incubation times, etc. Figure 1b shows a representative case with linear regression R=0.8119. For all 29 slides, linear regression R values ranged between 0.7750 and 0.9853, mean 0.8903. These graphs were used to normalize the individual raw AQUA scores from each field on the whole sections.
Inter-Slide Comparisons
Figure 2 shows slide-to-slide comparisons matched against the signout pathologist's score. Interslide differences appear to be minimized with the lowest scores. In our experience, AQUA ER scores greater than ten are typically considered positive associated with high pathologic scores and with better prognosis in TMA studies.14 Similarly, a pathologist's score of greater than 10% is traditionally considered positive. Using these a priori divisions to create binary values for AQUA and the pathologist's scores, the concordancy between AQUA and pathologist is reasonably good at 73% (κ score=0.526). Furthermore, the three discordant cases (F, G, and J) all show individual fields above the threshold for positive cases. If these field were selected (instead of the average) the concordance would be 100%. Table 1 shows normalized, mean AQUA scores for the different blocks. Again, using the binary values for AQUA ER scores, the concordancy between different blocks from the same case is 75% (24 out of a total of 32 block-to-block comparisons concordant). Of the comparisons that were considered discordant, the absolute differences were small (eg Case J/Block 1 vs Case J/Block 2 had AQUA scores of 10.151 and 7.035, respectively) (Table 1).
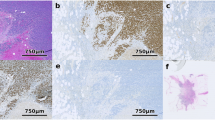
Most notable, however, are the discordancies between the pathologist and AQUA for cases F, G, J, and to a lesser degree case I (Figure 2 and Table 1). We attempted to retrieve the original ER slides assessed by the signout pathologist for these cases, but owing to the age of the research cases, we were only able to locate case I. Our review of this slide showed that the tumor appeared to be diffusely but weakly stained in a fairly nonspecific pattern (Figure 3a). There were however scattered areas of strong nuclear staining in adjacent benign components (Figure 3b). Because AQUA gives the average signal intensity in all pixels in a molecularly defined compartment (ie nucleus), it is possible that this may have accounted for the discordancy in this particular case. This case highlights the advantages of quantitative analysis when coupled to molecular compartment colocalization.
Because AQUA gives us continuous scores, we then performed unpaired t-tests and ANOVA analysis between the normalized, mean AQUA scores from different slides within individual cases (Table 1). Contrary to our results using binary divisions, this showed that only 6 of 32 (19%) slide-to-slide comparisons were concordant (t-test comparison P-value >0.05). Only cases A and I, 2 of 9 (22%), were concordant (ANOVA P-value >0.05). Case B was discordant with all three slide-to-slide comparisons significantly different. However, the AQUA scores were all extremely low (<5) making these cases likely ‘ER negative,’ and the significance of this discordancy unclear. In addition, this decreased our concordancy with the pathologist's score. At first, this seems contrary to our findings on our master/triplicate GSA in which we saw good correlation between AQUA scores and pathologist's scores (Figure 1). However, the 0.6 mm diameter histospots on the GSA were carefully chosen as the most homogeneous appearing areas of the tumor with likely more homogeneous ER expression, and multiple blocks from these cases were not evaluated. For the 29 whole sections, many fields from each section and multiple blocks from each case were analyzed, thus maximizing apparent tumor heterogeneity. These data confirm the fact that field selection for scoring can dramatically change outcome.
2-D Spatial Heterogeneity
To further characterize heterogeneity within individual slides, we looked at the pattern of ER expression on each slide. Our normalized AQUA ER scores ranged from 2.959 to 174.672. Most of the slides with low AQUA scores (≤10) were relatively tightly clustered with minimal variance (Figure 4a). However, as the scores on a given slide increased, the variance generally increased (Figure 4a). This finding did not seem to be strictly related to the number of fields analyzed per slide as high variance was seen with high number of fields (Figure 4a, Case C) as well as with low number of fields (Figure 4a, Case K). Corresponding 2-D ‘heat maps’ were also generated based on the normalized AQUA scores (Figure 4b). The heat maps show the relative score of each field with respect to all of the other fields on the same slide. As is traditionally done for array illustration, increasing red intensity correlates with concentrations of ER above the mean and increasing green intensity correlates with degree below the mean. Although for most slides, high and low scores on a given section appeared to be randomly scattered and with a normal distribution throughout the tumor, several slides showed a clustered pattern (Figure 4b, Cases J and 4b, and C). Interestingly, this clustering was seen in low scoring ‘ER negative’ cases as well as in higher scoring ‘ER positive’ cases. Indeed, scattered as well as clustered patterns were seen even on different blocks from the same case (Figure 4b, Case C). Corresponding whole sections stained with hematoxylin and eosin are also shown (Figure 4c) to demonstrate the areas of invasive cancers analyzed and the non-evaluable areas (shown as white squares in Figure 4b), which in general were areas without any tissue, minimum ‘maskable’ tumor, and areas having predominantly stroma or necrosis.
Scatter plots and 2-D representations of whole sections. (a–c) Representative data from five matched whole sections. (a) scatter plots of normalized AQUA scores of estrogen receptor are shown. Each image obtained on the section is assigned a field number starting in the upper left corner of the slide, across the row and down to the next row in a serpentine pattern. (b) heat maps showing AQUA ER scores assembled into a ‘virtual slide’ are shown. The most intense red are the highest ER scores and the most intense green are the lowest ER scores. (c) Corresponding H&E-stained whole sections used to identify areas of invasive cancers.
Discussion
In this report, we have used a quantitative measure of ER expression, AQUA, to demonstrate significant block-to-block heterogeneity of IHC reaction. Our results most likely reflect both laboratory-based variability owing to lack of standardization of immunohistochemistry and true biological heterogeneity. These results raise several questions. Whereas the AQUA algorithm allows separation of epithelial cells from stroma with the keratin mask, contamination of the analysis with benign and/or in situ epithelial cells remain a possibility. These confounding factors were minimized because, as described above, analysis was limited as much as possible to previously identified areas of invasive cancer, and because adjacent in situ tumors typically show similar ER expression compared with the invasive component.15 In addition, a number of investigators have described more intense staining in the periphery of tumors compared with the center.16 This can be attributed in part to increased necrosis in the center (these areas are eliminated from AQUA analysis with a crop function) and fixation artifact. Some have reported this pattern more in surgical specimens than in matched core biopsies, suggesting that improper fixation of the central tumor in large specimens may play a role.16 Several of our slides such as in (Figure 4b), Cases A and J also suggest more intense staining in the periphery. Alternatively, many investigators have described a phenomenon in which tumor at the invasive front shows different morphologies and preferentially expresses certain proteins vs tumor at the trailing edge/center.17 Although this has been most extensively described for colorectal cancers, this has also been described in breast cancer in the so-called prairie fire pattern.18 Irrespective of the reasons for the heterogenous pattern of ER expression within individual slides when using quantitative assessments, it is not surprising that there is a significant block-to-block heterogeneity within cases when using similar quantitative measurements.
At first, our concordancy rate appears lower than previous studies looking at different assays for ER in the same tumor, ER assessments in matched core biopsies and surgical resections, and comparative ER levels in matched primary tumors and their metastases.16, 19, 20, 21 However, earlier studies that used biochemical assays such as the dextran-coated charcoal method showed remarkably similar levels of ER discordancy within tumors ranging from 17 to 40% when using a binary cutpoint for ER positivity (eg ≥10 fmol/mg).22, 23, 24 When the differences in the mean content were compared within tumors however, the discordancy rate was significantly higher. Our results with AQUA using binary divisions and then continuous measurements mimic closely the results seen in these prior studies. This suggests that our methodology may have the capacity to combine the practicality, morphologic information achievable with an in situ assay, and arguably superior predictive capacity of IHC assessment of ER measurements with the continuous readouts of ER levels analogous to a biochemical assay. This may also be important because higher levels of ER expression both by biochemical methods and by semiquantitative IHC readings (eg Allred score) have been associated with a greater likelihood of endocrine therapy response.7, 25 Ultimately, the value of an exact ER score may be seen in the context of other exact scores like PR) in distinguishing among various hormonal therapy options.
In the future, it is conceivable that other potential prognostic and predictive biomarkers may also rely more heavily on continuous readouts and more accurate assessments of total tumor heterogeneity. For example, if the division for optimally choosing patients for a given biologic therapy is between the highest expressers vs the high expressers (eg AQUA <150 vs >150), simply looking for evidence of minimal staining (none vs any) before choosing to give treatment (as is often done for ER and tamoxifen treatment) and more cautious assessment of tumor heterogeneity for that particular biomarker will be necessary.
In summary, using AQUA to quantify in situ ER expression on multiple blocks from different primary breast cancers, we demonstrate reasonable correlation using traditional binary divisions, although significant slide-to-slide tumor heterogeneity was seen in the majority of cases when continuous scores were analyzed. Heterogeneity was greatest for those with higher levels of expression. It is likely that tumor heterogeneity is highly dependent on biomarker analyzed and caution should be used when making IHC determinants of biomarker expression in a tumor with single slide assessments.
References
Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–10874.
Giltnane JM, Rimm DL . Technology insight: identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol 2004;1:104–111.
Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–847.
Anderson E, Clarke RB, Howell A . Estrogen responsiveness and control of normal human breast proliferation. J Mammary Gland Biol Neoplasia 1998;3:23–35.
Sommer S, Fuqua SA . Estrogen receptor and breast cancer. Semin Cancer Biol 2001;11:339–352.
Fisher B, Redmond C, Fisher ER, et al. Relative worth of estrogen or progesterone receptor and pathologic characteristics of differentiation as indicators of prognosis in node negative breast cancer patients: findings from National Surgical Adjuvant Breast and Bowel Project Protocol B-06. J Clin Oncol 1988;6:1076–1087.
Early Breast Cancer Trialists' Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet 1998;351:1451–1467.
Layfield LJ, Saria E, Mooney EE, et al. Tissue heterogeneity of immunohistochemically detected estrogen receptor. Implications for image analysis quantification. Am J Clin Pathol 1998;110:758–764.
Camp RL, Chung GG, Rimm DL . Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 2002;8:1323–1327.
McCabe A, Dolled-Filhart M, Camp RL, et al. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst 2005;97:1808–1815.
Camp RL, Dolled-Filhart M, King BL, et al. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res 2003;63:1445–1448.
Chung GG, Yoon HH, Zerkowski MP, et al. Vascular endothelial growth factor, FLT-1, and FLK-1 analysis in a pancreatic cancer tissue microarray. Cancer 2006;106:1677–1684.
Psyrri A, Yu Z, Weinberger PM, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res 2005;11:5856–5862.
Camp RL, Dolled-Filhart M, Rimm DL . X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252–7259.
Leong AS, Sormunen RT, Vinyuvat S, et al. Biologic markers in ductal carcinoma in situ and concurrent infiltrating carcinoma. A comparison of eight contemporary grading systems. Am J Clin Pathol 2001;115:709–718.
Douglas-Jones AG, Collett N, Morgan JM, et al. Comparison of core oestrogen receptor (ER) assay with excised tumour: intratumoral distribution of ER in breast carcinoma. J Clin Pathol 2001;54:951–955.
Ono M, Sakamoto M, Ino Y, et al. Cancer cell morphology at the invasive front and expression of cell adhesion-related carbohydrate in the primary lesion of patients with colorectal carcinoma with liver metastasis. Cancer 1996;78:1179–1186.
Kauppila S, Stenback F, Risteli J, et al. Aberrant type I and type III collagen gene expression in human breast cancer in vivo. J Pathol 1998;186:262–268.
Allred DC, Bustamante MA, Daniel CO, et al. Immunocytochemical analysis of estrogen receptors in human breast carcinomas. Evaluation of 130 cases and review of the literature regarding concordance with biochemical assay and clinical relevance. Arch Surg 1990;125:107–113.
Mann GB, Fahey VD, Feleppa F, et al. Reliance on hormone receptor assays of surgical specimens may compromise outcome in patients with breast cancer. J Clin Oncol 2005;23:5148–5154.
Spataro V, Price K, Goldhirsch A, et al. Sequential estrogen receptor determinations from primary breast cancer and at relapse: prognostic and therapeutic relevance. The International Breast Cancer Study Group (formerly Ludwig Group). Ann Oncol 1992;3:733–740.
Davis BW, Zava DT, Locher GW, et al. Receptor heterogeneity of human breast cancer as measured by multiple intratumoral assays of estrogen and progesterone receptor. Eur J Cancer Clin Oncol 1984;20:375–382.
Lluch A, Anton A, Carbonell F, et al. Intratumoral variations in cytoplasmic estrogen receptor levels in breast cancer. Breast Cancer Res Treat 1987;9:69–70.
Silfversward C, Skoog L, Humla S, et al. Intratumoral variation of cytoplasmic and nuclear estrogen receptor concentrations in human mammary carcinoma. Eur J Cancer 1980;16:59–65.
Ellis MJ, Coop A, Singh B, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1- and/or ErbB-2-positive, estrogen receptor-positive primary breast cancer: evidence from a phase III randomized trial. J Clin Oncol 2001;19:3808–3816.
Acknowledgements
GG Chung received support from the DOD Breast Cancer Idea Award, W81XWH-04-1-0277, Anna and Argall Hull YCC Translational Research Award, and the Susan G Komen Breast Cancer Foundation BCTR0504220. DLR received support from the Breast Cancer Alliance of Greenwich CT and NIH Grants NCI R21 CA100825, NIH R33 CA 106709, and NIH R33 CA 110511.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer
GG Chung serves as a consultant for HistoRx Inc. DL Rimm and RL Camp are founder, stockholder and consultant to HistoRx Inc.
Rights and permissions
About this article
Cite this article
Chung, G., Zerkowski, M., Ghosh, S. et al. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest 87, 662–669 (2007). https://doi.org/10.1038/labinvest.3700543
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/labinvest.3700543
Keywords
This article is cited by
-
Reproducibility and intratumoral heterogeneity of the PAM50 breast cancer assay
Breast Cancer Research and Treatment (2023)
-
Selection of Intensity Distribution Characteristics in the Color Channels of Fluorescent Images of Cancer Cells
Journal of Applied Spectroscopy (2019)
-
Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer
Modern Pathology (2018)
-
Estrogen receptor quantitative measures and breast cancer survival
Breast Cancer Research and Treatment (2017)
-
Discordance between original and central laboratories in ER and HER2 results in a diverse, population-based sample
Breast Cancer Research and Treatment (2017)