Abstract
ERBB2 (HER-2/neu) amplification and/or overexpression are associated with poor prognosis in node-positive breast carcinoma. Its prognostic value in node-negative cases and its predictive value for response to chemotherapy remain controversial. This may be related to the use of molecular methods, which are sensitive to dilution of tumor material by normal cells, or the use of nonstandardized immunohistochemistry (IHC) procedures, for the determination of the ERBB2 gene status. In addition, new therapeutic approaches that target the cells overexpressing ERBB2 are under development. These perspectives necessitate a reliable evaluation of the status of ERBB2 in individual tumors before the application of specific therapeutic strategies. Fluorescent in situ hybridization (FISH) and IHC allow the evaluation of the ERBB2 status specifically in tumor cells on archival material. We have analyzed a series of 100 invasive ductal breast carcinomas without lymph node invasion both by IHC, using the CB11 monoclonal antibody and a sensitive Auidin Biotin Complex (ABC) immunodetection system, and by FISH, using the Oncor Inform HER-2/neu (ERBB2) gene amplification detection system as reference technique. Complete concordance between the results of FISH and IHC was seen in 98% of the cases. ERBB2 amplification (more than four signals per nucleus) was observed in 12 of the 100 cases, and all but one showed an overexpression of the protein (membrane staining) by IHC. Conversely, ERBB2 expression was present in one case without gene amplification. In conclusion, ERBB2 overexpression detected by IHC is highly correlated to gene amplification detected by FISH. Thus, under standardized conditions, IHC is a reliable and economical test to assess the ERBB2 status in tumors. The use of FISH could be limited to the verification of the status of tumors displaying a weak membrane immunostaining.
Similar content being viewed by others
INTRODUCTION
The ERBB2 (HER2/neu) oncogene, a member of the type I growth factor receptor gene family, located on the long arm of chromosome 17, is found amplified and/or overexpressed in about 20 to 30% of primary breast carcinomas (1, 2). Several works suggest that this amplification/overexpression is associated with poor prognosis in breast carcinoma, especially in node positive cases (1, 2, 3, 4, 5; for review, see 6, 7) and that it could be a marker of reduced response to chemotherapy (3, 4, 8, 9; for review, 6, 7). Conversely, whereas some authors provided data suggesting that ERBB2 expression was a marker of preferential response to anthracycline-containing regimens (10, 11), others found no significant correlation (12, 13, 14). The value of ERBB2 amplification/overexpression as an independent prognostic factor in node-negative cases remains controversial (8, 15, 16; for review, see 6). In addition, new therapeutic approaches that target the cells overexpressing ERBB2, based on monoclonal antibodies (17, 18, 19) and on antisense oligonucleotides (20), are under development. These perspectives necessitate an accurate knowledge of the ERBB2 status in individual tumors before the application of specific therapeutic strategies. ERBB2 gene status has been assessed mainly by Southern or dot blot, and by the polymerase chain reaction. Its overexpression has been detected by immunohistochemistry (IHC) in the majority of the cases, and by Western and Northern blot (for review, 6, 7). The range of positivity found in the various studies seems to be rather large: 5 to 55% for the amplification, and 10 to 55% for the overexpression (6). Indeed, results of molecular methods may be affected by the dilution of tumor material by normal cells and by the determination of the threshold defining the existence of an amplification or an overexpression (2, 7, 21). Recent technical developments have made possible the detection of the ERBB2 gene in tissue sections, touch preparations, and fine-needle aspirates using fluorescent in situ hybridization (FISH) (16, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32). The most reliable method to routinely evaluate the status of ERBB2 in prospective and archival material remains to be determined. Both FISH and IHC allow the evaluation of the status of ERBB2 on a cell-by-cell basis in archival material Therefore, they should be regarded as methods of choice for this assay. However, correlative results of these two approaches on the same samples are not yet amply available (26, 27, 28, 29, 30, 31, 32). To further assess the correlation between amplification and expression of this gene, and to evaluate ERBB2 in node-negative cases, we studied a series of 100 node-negative breast carcinomas, using both FISH and IHC on tissue sections from paraffin-embedded blocks.
MATERIALS AND METHODS
Patient Material
The samples analyzed were obtained from patients with invasive ductal carcinoma without lymph node involvement, who had undergone primary surgery only. Clinicopathological data of the patients and tumor samples studied are reported in Table 1. Tumor material consisted of 100 paraffin-embedded specimens fixed in AFA (75% ethanol 100°/5% acetic acid/2% commercial formalin/18% deionized water). For each case, the histology of the sample analyzed was verified on a hydroxyethyl starch-stained preparation, and areas showing invasive carcinoma were identified. The status of ERBB2 was independently assayed by FISH and IHC on two consecutive 4-μm tissue sections.
Immunohistochemistry
After deparaffinization and rehydration of the sections, endogenous peroxidases were inhibited by a 30-min incubation in 0.3% H2O2 in deionized water. All further steps were performed using an automated system. Tissue sections were incubated with the monoclonal anti c-erbB-2 oncoprotein NCL-CB11 (Novocastra Laboratories, Newcastle-on-Tyne, UK), diluted 1/400, for 1 h at 20°C, without antigen retrieval. The CB11 antibody was selected among two other antibodies assayed, A485 (Dako, Carpinteria, CA, USA), and Tab250 (Zymed, San Francisco, CA, USA), for its higher sensitivity and specificity in our experimental conditions. Immunodetection was achieved using the Vectastain Elite ABC peroxidase mouse IgG kit (Vector Laboratories, Burlingame, CA, USA), following the instructions given by the supplier. Then, slides were incubated with 3,3′-diaminobenzidine for 5 min and counterstained with Mayer's hematoxylin. A positive control was included in each processed series of slides. Preparations were analyzed independently by two investigators. Under these conditions, normal epithelial cells were not stained, and thus represented an internal negative control. The percentage of invasive carcinomatous cells showing a membranous staining, and the intensity of the staining (absent, weak, or strong) was recorded. The analysis lead to classify the tumors into three groups: cases without detectable staining in tumor cells [i.e., no ERBB2 overexpression (0)]; cases showing weak staining in the majority (more than 60%) of tumor cells [i.e., weak ERBB2 overexpression (+)]; cases showing strong staining in the majority (more than 60%) of the tumor cells [i.e., strong ERBB2 overexpression (++)]. Cases with only a minimal percentage of positive tumor cells were not observed.
FISH
FISH experiments were carried out using the Oncor Inform HER-2/neu (ERBB2) gene amplification detection system (Ventana Medical Systems, Tucson, AZ, USA), following the instructions given by the supplier. Briefly, deparaffinized slides were first treated with a protein-digesting enzyme at 37°C for 40 min. The preparations were denatured in 70% formamide/2× standard saline citrate (SSC), pH 7, at 75°C for 8 min. The mixture containing a biotinylated HER-2/neu DNA probe was applied to the denatured preparations, and slides were incubated for 15 hours at 37°C. The slides were washed in 50% formamide/2× SSC, pH 7, at 43°C for 15 min, then in 2× SSC, pH 7, at 37°C, for 10 min. Detection was achieved using fluorescein-labeled avidin, then an antiavidin antibody, followed by a second layer of fluorescein-labeled avidin. The nuclei were counterstained using a 4,6-DiAmidino-2-pheylIndole (DAPI)/antifade solution. The slides were viewed with an epifluorescence Leica DMRB microscope fitted with DAPI and double-band DAPI/fluorescein isothiocyanate filters. Images were captured using a Quantics digital camera (Photometrics, Tucson, AZ, USA) and Quips FISH imaging software (Vysis, Downers Grove, IL, USA). The number of fluorescein signals was counted in 20 nuclei of invasive tumor cells, referring to the hydroxyethyl starch-staining when necessary, in two distant areas of the section. For each tumor, the mean number of signals per nucleus was determined. Tumors showing a score of more than four signals per nucleus were considered to have ERBB2 amplification. In two cases in which a discordance between the results of FISH and IHC had been observed, a dual color FISH of ERBB2 and centromere of chromosome 17 (PathVysion HER-2 kit, Vysis) was performed, according to the instructions of the supplier.
Statistical Analysis
The concordance between the two methods was assessed using the kappa test. The sensitivity and specificity of IHC was evaluated, using dichotomous values (+, −), taking FISH as reference method.
RESULTS
The majority of the patients analyzed (68%) were older than 50 years. The large majority of the tumors were less than 20 mm in diameter (61%), and SBR grade II (63%).
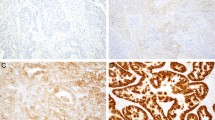
The results of the ERBB2 status as determined by FISH and IHC are given in Table 2, and representative pictures of FISH and IHC are shown in Figures 1 and 2, respectively.
FISH of ERBB2 probe on paraffin-embedded tissue sections of three cases of breast carcinoma. A, tumor without amplification. A mean number of two signals per nucleus is observed. B, tumor with a high level amplification. Presence of numerous signals, arranged in clusters. C, tumor with the lowest amplification (dual color FISH): presence of five to six signals of the ERBB2 probe (red) and two centromeres of chromosome 17 (green).
FISH
ERBB2 amplification was observed with the use of FISH in 12 of the 100 cases analyzed. In cases with an amplification, fluorescent signals were often arranged in clusters (Fig. 1B). In nine of these cases, the mean number of signals per nucleus was higher than 20. In the three other cases, the mean numbers were 12.5, 8.8, and 5.2. In this last case, the existence of an amplification was checked by dual color FISH with a centromeric probe of chromosome 17 in addition to the ERBB2 probe: tumor cells had a mean number of 1.7 signals corresponding to chromosome 17 (Fig. 1C).
Immunohistochemistry
ERBB2 overexpression was detected by IHC in 12 of the 100 cases analyzed. It was scored as strong (Fig. 2) in eight cases and weak in four cases. No discrepancies were observed between the two pathologists in the identification of positive cases. In our experimental conditions, the percentage of labeled tumor cells in positive cases always exceeded 60%, and the intensity of the staining could be readily scored according to one of the two defined categories (weak or strong). A concordance between amplification and detectable expression of the ERBB2 gene was found in 98 of the 100 cases examined (κ = 0, 90; P < 0.0001). Sensitivity of IHC, with FISH as the reference technique, was 92% (86 to 97%, 95% confidence interval), and specificity was 99% (97 to 100%, 95% confidence interval). Positive immunostaining was observed in 11 of the 12 cases with gene amplification. One case with weak ERBB2 expression showed no evidence of gene amplification. This case was checked by dual color FISH: the majority of tumor cells had two ERBB2 and chromosome 17 signals. Conversely, one case negative for ERBB2 expression by IHC had gene amplification (Table 2). This tumor showed a mean number of 8.8 ERBB2 gene copies per nucleus.
DISCUSSION
Reliable biological markers with prognostic and predictive significance are strongly needed for therapeutic decisions in breast carcinoma, in particular for the recognition of the 20 to 30% of lymph node-negative patients who will experience recurrence and for predicting the response to chemotherapy in node-positive patients. More than 10 years after the first report by Slamon et al. (1) showing the existence of a correlation between ERBB2 amplification and poor prognosis in breast carcinoma, the role of ERBB2 amplification/overexpression as an independent prognostic marker remains controversial, especially in node negative cases (3, 5, 8, 15, 16 for review, see 6, 7). Similarly, its predictive value regarding tumor response to treatment is still unclear (9, 10, 11, 12, 13, 14; for review, see 6, 7). A part of the uncertainty in establishing the prognostic and predictive value of ERBB2 may be related to technical difficulties in assessing the status of the gene by molecular techniques. In earlier studies, the status of ERBB2 was evaluated by Southern, Northern, or Western blot, and, more recently, by polymerase chain reaction. Results of these methods may be affected by the dilution of tumor material by normal cells (2, 21). These drawbacks have prompted the development of in situ techniques, FISH and IHC, which enable the recognition of the marker of interest in tumor cells only. Slamon et al. (2) have shown the existence of a direct correlation between amplification and overexpression of ERBB2. However, Press et al. (32) have stressed the variability of the sensitivity of HER-2/neu antibodies as potential source of errors in the assessment of the expression of the gene. The present controversy about the reliability of a U. S. Food and Drug Administration-approved HER-2/neu IHC test (33, 34) illustrates that this issue remains incompletely solved. Yet, the recently introduced therapeutic approaches, based on monoclonal antibodies (17, 18, 19) and on antisense oligonucleotides (20), that target tumor cells overexpressing ERBB2 require an accurate assessment of the status of the gene before inclusion of patients in such protocols.
These reasons mandate the necessity to calibrate IHC with the aid of a reference technique, which could be FISH. Farabegoli et al. (28), obtained a correlation of only 72% between FISH and IHC in a study of 79 primary invasive breast carcinoma samples, because of the observation of an excess of immunopositive cases without amplification. Similarly, Jimenez et al. (32) observed, in a comparative study of 41 cases, a proportion of 3 to 15% of cases, according to the antibody used, showing moderate immunostaining without amplification. Jacobs et al. (29), in a study of 100 consecutive paraffin-embedded breast carcinomas samples, demonstrated a concordance of 92% between the results of FISH and IHC, using the Oncor Inform kit and the Dako A485 polyclonal antibody. These results led the authors to recommend the use of IHC, under standardized conditions, as a routine technique for the evaluation of the ERBB2 status. Only one large study on node-negative breast carcinomas, using a combined approach by FISH and IHC on touch preparations has been reported (25). The authors used a HER-2/neu SpectrumOrange probe (Vysis) and a polyclonal anti-c-erbB-2 antibody (Dako). They observed a rate of 16% amplified cases and a concordance in 92% of the cases for ERBB2 gene amplification detected by FISH and protein overexpression detected by IHC.
We have observed a rate of 12% for ERBB2-amplified tumors in our series of node-negative breast carcinomas. Nine of the 12 tumors with ERBB2 amplification showed more than 20 copies of the gene per nucleus. Fluorescent signals were often arranged in clusters (Fig. 1B), corresponding to homogeneously staining regions observed in metaphase chromosomes of breast carcinomas (35). In our series, a concordance of 98% was observed between the gene status and its protein expression detected by IHC. One case with amplification did not display protein overexpression by IHC. Interestingly, this case showed a low mean number of signals per nucleus (8.8 per nucleus). Similar finding has been noticed by Jacobs et al. (29). The existence of cases with low-level amplification leads us to recommend the use of dual-color FISH with a centromeric probe of chromosome 17 to distinguish polysomy 17 from true ERBB2 amplification. In one case, a weak overexpression was observed without gene amplification. Comparing the results of the two techniques shows that if membrane labeling is present in a cluster of tumor cells, at any intensity, a gene amplification may exist. However, some rare cases with low intensity staining (+) may be unamplified. Such expression may be related to other mechanisms of activation of the ERBB2 gene (2, 29).
In our experience, the factors that optimally determined the results of IHC and gave a good correlation with FISH were the use of an automated protocol, the selection of the monoclonal antibody CB11, its appropriate concentration to yield no staining in normal epithelial cells, and the use of a sensitive ABC detection system. In conclusion, this study demonstrates that, IHC can reliably detect ERBB2-amplified tumors if rigorous conditions of sensitivity and standardization are applied. The use of FISH could therefore be limited to the control of the rare cases showing low protein expression by IHC.
References
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL . Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–182.
Slamon DJ, Goldophin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244: 707–712.
Gusterson BA, Gelber RD, Goldhirsch A, Price KN, Save-Soderborgh J, Anbazhagan R, et al. Prognostic importance of c-erbB-2 expression in breast cancer. International (Ludwig) Breast Cancer Study Group. J Clin Oncol 1992; 10: 1049–1056.
Têtu B, Brisson J . Prognostic significance of HER-2/neu oncoprotein expression in node- positive breast cancer. The influence of the pattern of immunostaining and adjuvant therapy. Cancer 1994; 73: 2359–2365.
Quénel N, Wafflart J, Bonichon F, de Mascarel I, Trojani M, Durand M, et al. The prognostic value of c-erbB2 in primary breast carcinomas: a study on 942 cases. Breast Cancer Res Treat 1995; 35: 283–291.
Révillion F, Bonneterre J, Peyrat J . ERBB2 Oncogene in human breast cancer and its clinical significance. Eur J Cancer 1998; 34: 791–808.
Ross JS, Fletcher JA . HER-2/neu (c-erbB-2) gene and protein in breast cancer. J Clin Pathol 1999; 112 (Suppl. 1): S53–S67.
Allred DC, Clark GM, Tandon AK, Molina R, Tormey DC, Osborne CK, et al. HER-2/neu in node-negative breast cancer: prognostic significance of overexpression influenced by the presence of in situ carcinoma. J Clin Oncol 1992; 10: 599–605.
Têtu B, Brisson J, Plante V, Bernard P . p53 and c-erbB-2 as markers of resistance to adjuvant chemotherapy in breast cancer. Mod Pathol 1998; 11: 823–830.
Muss H, Thor A, Berry D, Kute T, Liu ET, Koerner F, et al. c-erbB-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med 1994; 330: 1260–1266.
Paik S, Bryant J, Park C, Fisher B, Tan-Chiu E, Hyams D, et al. erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst 1998; 90: 1361–1370.
Rozan S, Vincent-Salomon A, Zafrani B, Validire P, De Cremoux P, Bernoux A, et al. No significant predictive value of c-erbB-2 or p53 expression regarding sensitivity to primary chemotherapy or radiotherapy in breast cancer. Int J Cancer (Pred Oncol) 1998; 79: 27–33.
Thor AD, Berry DA, Budman DR, Muss HB, Kute T, Henderson IC, et al. erbB-2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. J Natl Cancer Inst 1998; 90: 1346–1360.
Vincent-Salomon A, Carton M, Freneaux P, Palangie T, Beuzeboc P, Mouret E, et al. ERBB2 overexpression in breast carcinomas: no positive correlation with complete pathological response to preoperative high-dose anthracycline-based chemotherapy. Eur J Cancer 2000; 36: 586–591.
Rosen PP, Lesser ML, Arroyo CD, Cranor M, Borgen P, Norton L . Immunohistochemical detection of HER2/neu in patients with axillary lymph node negative breast carcinoma. A study of epidemiologic risk factors, histologic features, and prognosis. Cancer 1995; 75: 1320–1376.
Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, et al. Her-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 1997; 15: 2894–2904.
Baselga J, Norton L, Albanell J, Kim YM, Mendelsohn J . Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res 1998; 58: 2825–2831.
Pegram M, Lipton A, Hayes D, Weber BL, Baselga JM, Tripathy D, et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol 1998; 16: 2659–2671.
Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 1999; 17: 2639–2648.
Roh H, Pippin J, Boswell C, Drebin JA . Antisense oligonucleotides specific for the HER2/neu oncogene inhibit the growth of human breast carcinoma cells that overexpress HER2/neu. J Surg Res 1998; 77: 85–90.
de Cremoux P, Martin EC, Vincent-Salomon A, Dieras V, Barbaroux C, Liva S, et al. Quantitative PCR analysis of c-erb-B-2 (HER2/neu) gene amplification and comparison with p185(HER2/neu) protein expression in breast cancer drill biopsies. Int J Cancer 1999; 83: 157–161.
Kallioniemi O, Kallioniemi A, Kurisu W, Thor A, Chen LC, Smith HS, et al. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci U S A 1992; 89: 5321–5325.
Press MF, Hung G, Godolphin W, Slamon DJ . Sensitivity of Her-2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 1994; 54: 2771–2777.
Smith KL, Robbins PD, Dawkins HJ, Papadimitriou JM, Redmond SL, Carrello S, et al. c-erbB-2 amplification in breast cancer: detection in formalin-fixed, paraffin-embedded tissue by in situ hybridization. Hum Pathol 1994; 25: 413–418.
Pauletti G, Godolphin W, Press M, Slamon DJ . Detection and quantification of HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene 1996; 13: 63–72.
Persons DL, Borelli KA, Hsu PH . Quantitation of HER-2/neu and c-myc gene amplification in breast carcinoma using fluorescence in situ hybridization. Mod Pathol 1997; 10: 720–727.
Ratcliffe N, Wells W, Wheeler K, Memoli V . The combination of in situ hybridization and immunohistochemical analysis: an evaluation of Her2/neu expression in paraffin-embedded breast carcinomas and adjacent normal-appearing breast epithelium. Mod Pathol 1997; 10: 1247–1252.
Farabegoli F, Ceccarelli C, Santini D, Baldini N, Taffurelli M, Marrano D, et al. c-erbB-2 over-expression in amplified and non-amplified breast carcinoma samples. Int J Cancer 1999; 84: 273–277.
Jacobs T, Gown A, Yaziji H, Barnes MJ, Schnitt SJ . Comparison of fluorescence in situ hybridization and immunohistochemistry for evaluation of HER-2/neu in breast cancer. J Clin Oncol 1999; 17: 1974–1982.
Klijanienko J, Couturier J, Galut M, El-Naggar AK, Maciorowski Z, Padoy E, et al. Detection and quantification by FISH and image analysis of Her-2/neu amplification in breast cancer fine-needle samples. Cancer (Cancer Cytopathol) 1999; 87: 312–318.
Mezzelani A, Alasio L, Bartoli C, Bonora MG, Pierotti MA, Rilke F, et al. c-erbB2/neu gene and chromosome 17 analysis in breast cancer by FISH on archival cytological fine-needle aspirates. Br J Cancer 1999; 80: 519–525.
Jimenez RE, Wallis T, Tabasczka P, Visscher DW . Determination of Her-2/Neu status in breast carcinoma: comparative analysis of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol 2000; 13: 37–45.
Jacobs TW, Gown AM, Yasiji H, Barnes MJ, Schnitt SJ . Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drugs Administration-approved scoring system. J Clin Oncol 1999; 17: 1983–1987.
Roche PC, Ingle JN . Increased HER2 with U. S. Food and Drug Administration-approved antibody. J Clin Oncol 1999; 17: 434–435.
Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B . Detection of DNA amplification in 17 primary breast carcinomas with homogeneously staining regions by a modified comparative genomic hybridization technique. Genes Chromosom Cancer 1994; 10: 160–170.
Acknowledgements
The authors thank Dr. A. Nahal for thorough revision of the manuscript.
This work was supported in part by a grant of Oncor, Gaithersburg, MD.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Couturier, J., Vincent-Salomon, A., Nicolas, A. et al. Strong Correlation between Results of Fluorescent In Situ Hybridization and Immunohistochemistry for the Assessment of the ERBB2 (HER-2/neu) Gene Status in Breast Carcinoma. Mod Pathol 13, 1238–1243 (2000). https://doi.org/10.1038/modpathol.3880228
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/modpathol.3880228
Keywords
This article is cited by
-
Clinical role of HER2 gene amplification and chromosome 17: a study on 154 IHC-equivocal cases of invasive breast carcinoma patients
Tumor Biology (2016)
-
Prospective multi-centre study to validate chromogenic in situ hybridisation for the assessment of HER2 gene amplification in specimens from adjuvant and metastatic breast cancer patients
Journal of Cancer Research and Clinical Oncology (2011)
-
Current issues in ER and HER2 testing by IHC in breast cancer
Modern Pathology (2008)
-
High concordance between immunohistochemistry and fluorescence in situ hybridization testing for HER2 status in breast cancer requires a normalized IHC scoring system
Modern Pathology (2008)
-
Prognostic value of hormonal receptors, p53, ki67 and HER2/neu expression in epithelial ovarian carcinoma
Clinical and Translational Oncology (2008)